Abstract
We evaluated the effects of seed- and pollen-mediated gene dispersal on genetic structure among Quercus salicina saplings. Parentage analysis using 10 microsatellite markers indicated that the 111 adult trees located within a 11.56 ha plot in the Tatera Forest Reserve, Japan, included only one parent of 44.2% and both parents of 40.7% of the 226 saplings located in a 1-ha core plot at its center. Coancestry (Fij) estimates indicated that there was strong genetic structure among the saplings. The numbers of pairs of full- and half-siblings were high among neighboring saplings, suggesting that there was strong maternal half-sibling family structure among the saplings around their seed parents, probably generated by the spatially limited seed dispersal and the small extent of overlapping seed shadows owing to the low density of adults. The frequencies also suggest that the maternal half-sibling families are interspersed with full-siblings, produced by correlated mating, probably because pollination frequency depends on the distance between parents. The frequencies of pairs of half-siblings decreased as the distance between saplings increased, but did not fall to zero even at distances up to the 90–95 m class, suggesting that paternal half-siblings originating from correlated paternity were widely distributed owing to extensive pollen flow. We separately examined the genetic structure for maternal and paternal alleles in the saplings. Unsurprisingly, very strong genetic structure was detected for maternal alleles. However, weak (but significant) genetic structure was also detected for paternal alleles. Therefore, pollen dispersal may affect the extent of genetic structure as well as seed dispersal.
Similar content being viewed by others
Introduction
The genetic structure within plant populations is a consequence of the combined effects of various evolutionary forces—such as gene flow, genetic drift and selection—that are influenced by ecological factors including the mating system, the limitations of seed and pollen dispersal, adult density, microenvironmental variations and colonization (Epperson, 1993; Hamrick et al., 1993; Kalisz et al., 2001). Limited seed dispersal should affect both the existence and extent of genetic structure, whereas the other factors should affect only its extent. Most species of flowering plants are cosexual, producing either hermaphroditic flowers with both male and female parts or separate, monoecious male and female flowers (Silvertown and Charlesworth, 2001). Adults of such species function as both maternal and paternal parents, and thus their offspring constitute two kinds of half-sibling families (maternal and paternal), with varying frequencies of full-siblings. Seed dispersal is usually highly limited compared with pollen dispersal (Ennos, 1994). Therefore, the offspring tend to be spatially organized as aggregates of maternal half-siblings in patches around their maternal parents. The distribution of paternal half-sibling families is influenced by both the pollen dispersal and the seed dispersal of the maternal parents that accept the pollen. Therefore, the distribution of maternal half-sibling families is generally more restricted than that of paternal half-sibling families, especially for species with long-distance pollen flow.
Hardy et al. (2004) found that about a one-fifth of the offspring pairs within maternal progeny arrays of Centaurea corymbosa they investigated were full-siblings, and that the extent of correlated paternity (the proportion of offspring sired by the same paternal parent) in different maternal families decreased with the distance between maternal parents. They concluded that spatially limited pollen dispersal was a major contributory factor to the correlated paternity. When seed dispersal is limited, saplings should aggregate around each seed parent, producing spatial family structures of maternal half-siblings. In addition, if pollen dispersal is also limited, correlated mating (correlated paternity within the same maternal progeny arrays) can occur. Full-siblings should then appear in the aggregates of maternal half-siblings around the respective seed parents. The probability of pairs of randomly sampled saplings from different maternal half-sibling families being paternal half-siblings may decrease with the distance between the maternal half-sibling families. Because of the limitations of seed and pollen dispersals, there should be strong genetic structure in sapling populations. Recently, the development of microsatellite markers, which are codominantly expressed and tend to have very high polymorphism, has greatly facilitated parentage analysis in plant populations (Chase et al., 1996; Dow and Ashley, 1996, 1998; Streiff et al., 1999; Isagi et al., 2000). Such parentage analysis could allow us to assign maternal and paternal parents to saplings, to identify saplings with half-, full- and non-sibling relationships, to distinguish between maternal and paternal alleles in the genotypes of saplings and to analyze the spatial distribution of paternally and maternally derived alleles. The results obtained could be very useful for testing hypotheses regarding genetic structure, such as the one postulated above.
The species studied here, Quercus salicina, is a wind-pollinated tree species, with gravity-dispersed seeds. In a previous study (Nakanishi et al., 2004) of the species, the pollen pools accepted by seed parents were found to differ genetically among the seed parents, and the extent of differentiation was positively correlated with the spatial distance between the seed parents. The results may reflect distance-dependent correlated paternity because of the distance dependence of pollen flow at a local scale in this species. The seed dispersal of this species should be strongly limited because its seeds are primarily dispersed by gravity, and thus saplings tend to establish around their seed parents. Therefore, family structure may develop, with aggregations of half-siblings interspersed with full-siblings, around seed trees, and thus spatial genetic structure not only of maternal alleles but also of paternal alleles may appear. In this study, we evaluated the effects of seed- and pollen-mediated gene dispersal on genetic structure among Q. salicina saplings. The evaluations were conducted through (1) parentage analysis to assign parents of saplings and identify half-, full- and non-sibling relationships among them using microsatellite markers; (2) analyses of the spatial genetic structure of saplings; and (3) analyses of the genetic structure of maternal and paternal alleles in the saplings.
Materials and methods
Study site and field methods
The study site was in the Tatera Forest Reserve, on the South Island of Tsushima, which is located between the Japanese Archipelago and the Korean Peninsula. The reserve, protected as a National Natural Monument, has an area of approximately 100 ha and is situated on the north-facing slope of Mt Tatera. There has been no human interference in the reserve for several centuries, and an old-growth evergreen broad-leaved forest is well developed in it (Itow, 1991).
A 4-ha permanent plot (200 × 200 m) was established in this reserve in 1990. Tree censuses have been conducted for all stems ⩾5 cm in diameter at breast height (d.b.h.) of all woody plant species, and the vegetational features of the plot have been described by Manabe et al. (2000). In 2000, a 1-ha core plot (100 × 100 m) was established in the center of the 4-ha plot, and a census was performed within the 1-ha core plot, in which 247 Q. salicina individuals ⩾30 cm in height and with a d.b.h. of <5 cm (defined as saplings in this study) were observed and mapped. In 2003, the 4-ha plot was expanded to 11.56 ha (340 × 340 m) for a census of Q. salicina, and 111 Q. salicina individuals were found with a d.b.h. of ⩾5 cm (Figure 1). Our previous field observations of Q. salicina in the plot indicated that all individuals with a d.b.h. of ⩾5 cm can flower, but that individuals with a d.b.h. of <5 cm are not yet ready to flower. Thus, all the 111 individuals could be defined as adult trees with the potential to act as parents. Leaf samples from all 111 adults were collected in 1999 and 2003, and from all 247 saplings in 2000. The collected leaf samples were transported to our laboratory and stored at −30 °C until DNA extraction.
DNA extraction and microsatellite genotyping
The methods used for DNA extraction and microsatellite genotyping in this study were the same as those used in our previous study (Nakanishi et al., 2004). Genotype data for all adults and saplings of Q. salicina were collected for 10 microsatellite loci: QsalCT15 and QsalCT33 developed for Q. salicina by T Kawahara of Hokkaido Research Center, Forestry and Forest Products Research Institute, Japan (Nakanishi et al., 2004), QpZAG36 and QpZAG119 (Steinkellner et al., 1997), MSQ4 (Dow et al., 1995), QrZAG7, QrZAG20, QrZAG25 and QrZAG101 (Kampfer et al., 1998), and QM69-2M1 (Isagi and Suhandono, 1997). The genotypes of the adults at seven loci (QsalCT15, QsalCT33, QpZAG119, MSQ4, QrZAG7, QrZAG101 and QM69-2M1) had already been determined (Nakanishi et al., 2004). The genotypes of adults at the remaining 3 loci, as well as the genotypes of saplings at all 10 loci, were analyzed in this study.
Statistical data analyses
The genetic diversity of the Q. salicina adults and saplings was estimated using standard population genetic parameters calculated by Genepop version 3.4 (Raymond and Rousset, 1995). The number of alleles (A), allelic richness (R), observed heterozygosity (HO), gene diversity (HE) and fixation index (FIS) were calculated for each locus and over all loci. The deviations from the Hardy–Weinberg equilibrium (HWE) at each locus and over all loci were tested by the Markov chain method.
Parentage analysis was performed by the maximum likelihood method based on multilocus genotypes (Meagher and Thompson, 1986) of all the adults and saplings, using FaMoz (Gerber et al., 2003), following the method by Gerber et al. (2000). The most likely parents and parent pairs were determined by means of log of the odds ratio (LOD) scores using the reference allele frequencies, which were calculated in the adult population within the 11.56 Ha plot. The test thresholds of LOD scores for rejecting a candidate parent as a true parent (TP), and for rejecting a candidate parent pair as the true one (TC) were determined by the simulation procedure (see details in Gerber et al., 2000, 2003). At the same time as the simulation procedure, we also calculated the probabilities of correct classification (the most likely parent or parent pair being the true parent or parent pair), and type I errors (erroneous rejection of the null hypothesis that the parents are present inside the plot) and type II errors (erroneous acceptance of the null hypothesis) at the TP and TC. We incorporated an error rate at genotyping of 0.1%, as used in previous studies (Gerber et al., 2000; Sezen et al., 2005; Setsuko et al., 2007), into both of the above-mentioned calculations of LOD scores and simulations. If a candidate had an LOD score exceeding TP, it was considered a true potential parent, and if a parent pair had an LOD score greater than TC and comprised two true potential parents, it was considered a true potential parent pair (Meagher and Thompson, 1986). If a sapling had no true potential parent, it was regarded as having no parent within the plot. If a sapling had only one true potential parent or parent pair, the potential parent or parent pair was regarded as its true parent or parent pair. If a sapling had multiple potential parents or parent pairs, it was regarded as having a parent or parent pair within the plot, but its parentage could not be assigned.
To examine the spatial family structure of saplings (spatial aggregation of half- and full-siblings), we calculated the numbers of pairs of saplings at each distance class, being full-, half- and non-siblings. The distance classes were defined at continuous 5-m intervals. The analysis was conducted among the saplings that had unambiguously assigned parent pairs after the parentage analysis.
The spatial genetic structure of saplings in the 1-ha core plot was evaluated by calculating Fij (coancestry; Loiselle et al., 1995) kinship coefficients. A kinship coefficient is generally defined as the probability that the compared genes are identical by descent (e.g. Ritland, 1996), but Fij actually estimates relative kinship, which can be defined as the ratio of differences of probabilities of identity in state (Rousset, 2002). In the case of two individuals i and j, Fij between them can be defined as Fij=(Qij−Qm)/(1−Qm), where Qij is the probability of identity in state for random genes from i and j, and Qm is the average probability of identity by state for genes coming from random individuals from the sample (or ‘reference population’). When the reference population is the parental generation, Fij values should theoretically be identical to values predicted from pedigree information and be 0.125 for half-siblings and 0.250 for full-siblings (Hardy and Vekemans, 2002). For this analysis, we used the parental generation composed of all 111 adult trees in the 11.56-ha plot as the reference population (that is, the ‘reference allele frequencies’ used in the parentage analysis mentioned earlier). The average Fij value was calculated for each of 20 continuous distance classes at 5-m intervals from 0–5 to 95–100 m. The significance of the average Fij values was then assessed by permutation tests (with 1000 permutations), in which spatial distances were permuted randomly among pairs of saplings. These calculations were performed by SPAGeDi version 1.2 (Hardy and Vekemans, 2002), except that the permutation tests were conducted manually. We also calculated the average Fij value between parents (seed parents or pollen parents) and their respective saplings and those of full-, half- and non-siblings among the saplings for which parents offspring and sibling relationships could be determined by parentage analysis, and compared the observed values with the expected values (0.250, 0.250, 0.125 and 0.000 for the relationships of parents offspring, full-, half- and non-siblings, respectively) to evaluate the validity of the calculation using the reference allele frequencies.
To determine the seed parents and pollen parents of saplings, we adopted two assumptions by Dow and Ashley (1996) and Bacles et al. (2006) as follows: (i) if a sapling had only one parent within the 11.56-ha plot, this parent was assumed to be the seed parent; and (ii) if a sapling had a parent pair within the plot, the closer parent was assumed to be the seed parent and the more distant parent to be the pollen parent. These two assumptions are based on the fact that seeds of the Quercus species generally disperse over shorter distances than the pollen because the seeds and pollen are primarily dispersed by gravity and wind, respectively. Therefore, seed parents should generally be closer to their offspring than pollen parents. In addition, we assumed that none of the parents within the plot had died. After determining seed parents and pollen parents, we compared the average values of the spatial distances between saplings and their seed and pollen parents with the pollination distance using the Mann–Whitney U-test to roughly show the validity of the assumption that the closest identified parent was the seed parent and the more distant parent was the pollen parent. If the assumption is valid, the average distance between saplings and their more distant putative pollen parents should not be different from the average pollination distance, but that between saplings and their closest putative seed parents should be shorter.
Following the methods published by Hardy et al. (2004), we also examined spatial genetic structure among haplotypes derived from seed and pollen parents, separately, of saplings for which one parent or parent pair had been unambiguously determined. The paternal allele of each sapling was identified by subtracting the maternal allele from the sapling genotype (Smouse et al., 2001; Hardy et al., 2004), and then the maternal and paternal haplotypes were expediently converted into diploid homozygous genotypes with the alleles to calculate Fij values. In ambiguous cases, in which both the offspring and its respective seed parent were heterozygotes with the same alleles, the maternal and paternal haplotypes were expediently converted into the corresponding heterozygous genotypes. The spatial genetic structure of the paternal and maternal haplotypes of saplings was then examined in the same way that the spatial genetic structure of the saplings' genotypes was examined.
Results
Genetic diversity in adults and saplings
Genotypes of most individuals (108 of the 111 adults within the 11.56-ha plot and 193 of the 247 saplings within the 1-ha core plot) could be determined at all 10 of the loci examined. Genotype data were missing regarding 1, 2 or 3 loci for the other three adults, and thus 0.5% of the 1110 genotypes at the 10 loci among the 111 adults were missing in total. There were also missing genotype data for saplings. In the following analyses, we used 226 saplings, the genotypes of which could be determined at more than five loci. Of the 2260 genotypes, 96 over the 10 loci among the 226 saplings (4.2%) were missed. However, both the FaMoz and SPAGeDi programs could take account of the missing data.
The 10 loci were highly polymorphic in both adults and saplings, with 7–39 and 4–42 alleles per locus and mean HE values of 0.783 and 0.728, respectively (Table 1). FIS values for each locus ranged from −0.117 to 0.087 and −0.091 to 0.144, and FIS values for all the loci were 0.006 and −0.009 for adults and saplings, respectively. Deviations from HWE were not significant at any of the loci in adults, but there was significant deviation at the QrZag20 locus in saplings (P<0.001 after Bonferroni correction).
The total exclusion probabilities calculated for reference allele frequencies were 0.999 and 1.000 for single parents and parent pairs, respectively, over the 10 loci. For all the loci, frequencies of null alleles were lower than 0.05.
Parentage and spatial family structure of saplings
From the results of the simulation procedure, we determined TP and TC values to be 4.2 and 12.4, respectively. Estimated probabilities of correct classifications were 95.8 and 99.5% for single parent and parent-pair assignments, respectively. At the TP, the estimated probabilities of type I and type II errors for single parent assignments were <0.06 and <0.12, respectively, whereas at the TC, the corresponding probabilities for parent-pair assignments were <0.01 and 0.00, respectively.
Of the 226 saplings, 34 (15.0%) had no true potential parent in the 11.56-ha plot. The other 192 (85.0%) saplings had at least one true potential parent in the plot and were defined as having at least one parent in it (Table 2). Of the 192 saplings, 92 (47.9%) had at least one true potential parent pair in the plot and were defined as having both parents in the plot, whereas the other 100 (52.1%) had no true potential parent pair in it. Of these 100 saplings, 85 had only one true potential parent, which could thus be defined as the true parent (unambiguous assignment for single parent), and 15 had multiple true potential parents (ambiguous assignment for single parent). Of the 92 saplings that had at least one true potential parent pair, 83 had only one such pair, which could thus be defined as the true parent pair (unambiguous assignment for a parent pair), whereas 9 saplings had multiple true potential parent pairs (ambiguous assignment for parent pair). Assuming that no parents in the 11.56-ha plot had died, one or both parents of 44.2% (100 saplings) and 15.0% (34) of the 226 saplings were located outside the 11.56 ha plot, respectively. Thus, the rate of seed flow from outside the plot should theoretically be 15.0%. Among the 192 saplings that had at least one parent within the plot, 52.1% (100) apparently had one parent outside the plot, and this percentage should theoretically mirror the rate of pollen flow from outside the plot. The analyses of the 226 saplings indicated that 43 (38.7%) of the 111 potential parents within the plot were parents of at least one sapling, with the numbers of offspring apparently ranging from 1 to 59 (5.8±10.0 on average). Here and elsewhere in the text, average and error values refer to means±s.d.
The following results were derived from analyses of the 83 saplings with unambiguously assigned parent pairs. The average distance between saplings and their closest parents (putative seed parents) was 16.8±17.0 m, significantly shorter than the average pollination distance (69.2±52.5 m) (Mann–Whitney U-test, P<0.001), but the average distance between saplings and their more distant parents (putative pollen parents) was 74.4±45.8 m and did not differ significantly from the average pollination distance (Mann–Whitney U-test). The saplings appeared to be aggregated around their closest parents, which were probably their seed parents in most cases (Figure 2). Eleven and 33 different adults within the plot were found to be the putative seed and pollen parents of at least one sapling, respectively. The number of offspring of each putative seed and pollen parent ranged from 1 to 36 and 1 to 15, with averages of 7.5±10.5 and 2.5±3.1, respectively. The average distances between pairs of full-, half- and non-sibling saplings were 15.5±9.6, 20.1±17.6 and 48.1±22.4 m, respectively (Figure 3). The frequencies of pairs that were full- and half-siblings were much higher in short distance classes than in long distance classes; 114 (91.2%) and 668 (74.5%) of all the full- and half-sibling pairs were detected in the 0–5 to 20–25 m classes, respectively. The number of full-siblings decreased rapidly at 25–30 m and fell to zero in the 40–45 m and longer distance classes (except in the 70–75 m distance class), but the number of half-siblings decreased gradually as the distance increased, and did not fall to zero until the 90–95 m distance class (Figure 3).
Location of saplings and their closer parents (putative seed parents), which produced more than 10 saplings within the 1-ha core plot. Large letters represent the parents of the saplings indicated by the same small letters. Large and small filled circles represent the other adults within and around the 1-ha core plot and saplings within the core plot, respectively.
Spatial genetic structure of saplings
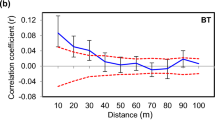
The following results were derived from the analyses of all 226 saplings. Significantly positive Fij values were obtained for the 0–5 to 20–25 m distance classes according to the permutation test (P<0.05), and the value was highest (0.0954) for the shortest distance class (Figure 4). The Fij values decreased rapidly as distance increased and were significantly lower than the 95% confidence intervals (two-tailed) of Fij values in the 25–30 to 95–100 m distance classes. For all the distance classes analyzed, the Fij values were positive. The average Fij value between parents and their respective saplings and those of full-, half- and non-siblings were 0.233±0.056, 0.246±0.079, 0.115±0.078 and 0.015±0.063, respectively. The observed average values were approximately equivalent to the expected values.
Correlogram of average Fij values for saplings calculated using the reference allele frequencies. Distance classes were defined at continuous 5-m intervals from 0–5 to 95–100 m. The dashed lines represent the 95% (two-tailed) confidence interval of the average Fij distribution calculated by 1000 permutations of spatial distances among pairs of saplings.
Spatial genetic structure of maternal and paternal alleles in saplings
The following results were derived from the analyses of the 168 saplings that had unambiguously assigned parents or parent pairs. Significantly positive Fij values were detected in the 0–5 to 20–25 and 0–5 to 15–20 distance classes for maternal and paternal alleles, respectively (P<0.05, according to the permutation test), and the highest values for maternal and paternal alleles were in the first and third distance classes, respectively (0.275 and 0.046, respectively; Figure 5). The Fij values of the maternal and paternal alleles decreased rapidly and gradually as the distance classes increased, respectively, but nevertheless were positive for almost all of the distance classes. The Fij values for particularly short-distance classes were much higher for the maternal alleles than for the paternal alleles.
Correlograms of average Fij values for maternal and paternal alleles of saplings calculated using the reference allele frequencies. Distance classes were defined at continuous 5-m intervals from 0–5 to 95–100 m. The dashed lines represent the 95% (two-tailed) confidence interval of average Fij distribution calculated by 1000 permutations of spatial distances among pairs of saplings.
Discussion
Patterns of seed and pollen dispersal
Saplings appeared to be spatially aggregated around their closest parents. Furthermore, the average distance between saplings and their more distant putative parents was not significantly different from the average pollination distance, but the average distance between saplings and their closest putative parents was significantly shorter. These findings are consistent with the assumption that closer and more distant parents are seed and pollen parents, respectively, especially because the density of adults was low (9.6 ha−1) and they had a scattered distribution in the study population. Dow and Ashley (1996), who examined the seed dispersal of another Quercus species, Quercus macrocarpa, under the same assumptions as in this study, found clusters of half-siblings around one of their respective parents and concluded that the assumption seemed to be justified because the clustering pattern of the saplings was easily explained by limited seed dispersal, but it was difficult to envision a scenario in which paternally related acorns from different seed parents would come to be established around a common pollen donor. Our findings provide further empirical indications that the assumption is valid in most cases.
If the assumption is valid, the distances between saplings and their closest parents should correspond to the distances of seed dispersal. The average distance of seed dispersal in the study population of Q. salicina was estimated to be 16.8±17.0 m, which was similar to the corresponding distances for Q. macrocarpa (22.8±24.8 m) and Quercus pyrenaica (14 m) reported by Dow and Ashley (1996) and Valbuena-Carabana et al. (2005), respectively. The proportion of seed flow from outside the 11.56-ha plot was estimated to be 15.0%, and all the saplings were located at least 120 m from any edge of the 11.56-ha plot. Therefore, 15.0% of seed flow was estimated to occur over a distance greater than 120 m. Although most of the seed movements may occur over short distances by gravity, long-distance seed flow by animals may also occur with a low frequency. The result in this study is approximately consistent with those from studies of other Quercus species (Dow and Ashley, 1996; Valbuena-Carabana et al., 2005). However, the seed dispersal of Q. salicina was very limited when compared with those of species with seeds dispersed by animals (Sezen et al., 2005) and wind (Bacles et al., 2006).
As the proportion of pollen flow from outside the plot was 52.1%, the average distance of the total pollen flow should be greater than the estimated average distance of pollen flow within the plot (69.2 m). Therefore, long-distance pollen flow by wind may occur in the study population, as it does in other Quercus species (Dow and Ashley, 1996, 1998; Streiff et al., 1999) and other wind-pollinated species (Bittencourt and Sebbenn, 2007). Moreover, seed dispersal appears to be highly limited compared to pollen dispersal in Q. salicina, as reported for other Quercus species (Ennos, 1994; Dow and Ashley, 1996). The average distance of pollen flow within the plot (69.2±52.5 m) and proportion of pollen flow from outside the plot (52.1%), found in this study by parentage analysis using saplings, were very similar to those (66.7±40.0 m and 52.2%) found in our previous study (Nakanishi et al., 2004) based on paternity analysis using seeds of the same population at the same plot. The previous study found that the average distance of pollen flow within the plot was significantly shorter than that expected under random mating, and the frequency of pollination within the plot depended on the distance between the parents, prompting the conclusion that effective pollen dispersal in Q. salicina may be governed by a combination of two processes: local dispersion and long-distance transport, as advocated by Streiff et al. (1999) following their study of Q. robur L. and Q. petraea. The results in this study may reflect the pattern of pollen flow deduced from our previous seed-stage analysis (Nakanishi et al., 2004).
Spatial genetic structure and family structure of saplings
The correlogram of coancestry (Fij) between saplings indicated that there was a strong genetic structure among them. When adults in the 11.56 ha plot were used as the reference population, the average Fij value in the shortest distance class (0.0954) was high, and the numbers of pairs of saplings that were full- and half-siblings were relatively high in short-distance classes. These results indicate that there is a strong maternal half-sibling family structure among the saplings around seed parents, probably generated by the limited seed dispersal and small extent of overlapping seed shadows, owing to the low density of adults (9.6 per ha) at this study site, which is located in an old-growth evergreen broad-leaved forest. The numbers of half-siblings decreased as the distance increased, but did not fall to zero even in long-distance classes, indicating that paternal half-siblings are widely distributed, probably because of the correlated paternity (offspring from a given mother being sired by the same paternal parent) between different maternal progeny arrays, mediated by extensive pollen flow. Of all the full-sibling pairs, 114 (91.2%) were detected in distance classes from 0–5 to 20–25 m, indicating that the aggregates of maternal half-siblings around seed parents were interspersed with full-siblings. Hardy et al. (2004) reported that limited pollen dispersal was a major contributory factor to correlated mating (correlated paternity within maternal progeny arrays) in C. corymbosa. Thus, the full-siblings in the aggregates of half-siblings may be produced by correlated mating because of the inverse relationship between pollination frequency and the distance between parents at a local scale in Q. salicina. The aggregation of full-siblings around seed parents should strengthen the spatial genetic structure of saplings.
To examine whether the observed spatial genetic structure in the saplings is maintained until the adult stage, we re-analyzed the spatial genetic structure among the 111 adults based on Fij values estimated using the 10 loci (data not shown), as analyzed in this study. We confirmed significant spatial genetic structure among the adults, as observed in our previous study (Nakanishi et al., 2004), indicating that the genetic structure among the adults reflects that among the saplings. Our previous paternity analysis using seeds of the same population at the same plot (Nakanishi et al., 2004) suggests that the genetic structure among adults and limited pollen dispersal may produce biparental inbreeding (that is, nearby adult trees tend to be genetically related to seed parents because of the genetic structure, and may often be pollinated by the seed parents because of the short distance between them). The process may also strengthen the spatial genetic structure of saplings and partially explain the significant deviation from HWE for the QrZAG20 locus in saplings.
Genetic structure of maternal and paternal alleles in saplings
Unsurprisingly, a very strong genetic structure was detected for maternal alleles. However, a significant (albeit weak) genetic structure was also detected for paternal alleles. The genetic structure of the paternal alleles was probably because of correlated mating, mediated by limited pollen dispersal, generating full-siblings in the aggregates of half-siblings around the respective seed parents. In the correlogram of paternal alleles, the Fij values decreased more gradually as distance increased than in the corresponding correlogram of maternal alleles. The differences presumably reflect the differences between seed- and pollen-mediated gene dispersal resulting from the average pollination distance being greater than the average seed dispersal distance, and thus paternal half-siblings generally being more widely dispersed than maternal half-siblings. The genetic structures of the maternal and paternal alleles indicate that limited seed dispersal is primarily responsible for generating strong genetic structure among Q. salicina saplings, but limited pollen dispersal strengthens it.
In summary, we evaluated the effects of gene dispersal through seeds and pollen on genetic structure among Q. salicina saplings by reconstructing the dispersal patterns of the seed and pollen, and analyzing the consequent spatial genetic structure and family structure of the saplings as well as the genetic structure of their maternal and paternal alleles. The results suggest that the limited seed dispersal is primarily responsible for generating strong genetic structure among these saplings, but pollen flow contributes to it through correlated mating.
References
Bacles CFE, Lowe AJ, Ennos RA (2006). Effective seed dispersal across a fragmented landscape. Science 311: 628.
Bittencourt JVM, Sebbenn AM (2007). Patterns of pollen and seed dispersal in a small, fragmented population of the wind-pollinated tree Araucaria angustifolia in southern Brazil. Heredity 99: 580–591.
Chase MR, Moller C, Kesseli R, Bawa KS (1996). Distant gene flow in tropical trees. Nature 383: 398–399.
Dow BD, Ashley MV (1996). Microsatellite analysis of seed dispersal and parentage of saplings in bur oak, Quercus macrocarpa. Mol Ecol 5: 615–627.
Dow BD, Ashley MV (1998). High levels of gene flow in bur oak revealed by paternity analysis using microsatellites. J Hered 89: 62–70.
Dow BD, Ashley MV, Howe HF (1995). Characterization of highly variable (GA/CT)n microsatellites in the bur oak, Quercus macrocarpa. Theor Appl Genet 91: 137–141.
Ennos RA (1994). Estimating the relative rates of pollen and seed migration among plant populations. Heredity 72: 250–259.
Epperson BK (1993). Recent advances in correlation studies of spatial patterns of genetic variation. Evol Biol 27: 95–155.
Gerber S, Chabrier P, Kremer A (2003). FAMOZ: a software for parentage analysis using dominant, codominant and uniparentally inherited markers. Mol Ecol Notes 3: 479–481.
Gerber S, Mariette S, Streiff R, Bodenes C, Kremer A (2000). Comparison of microsatellites and amplified fragment length polymorphism markers for parentage analysis. Mol Ecol 9: 1037–1048.
Hamrick JL, Murawski DA, Nason JD (1993). The influence of seed dispersal mechanisms on the genetic structure of tropical tree populations. Vegetatio 107/108: 281–297.
Hardy OJ, Gonzalez-Martinez SC, Colas B, Freville H, Mignot A, Olivieri I (2004). Fine-scale genetic structure and gene dispersal in Centaurea corymbosa (Asteraceae). II. Correlated paternity within and among sibships. Genetics 168: 1601–1614.
Hardy OJ, Vekemans X (2002). SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2: 618–620.
Isagi Y, Kanazashi T, Suzuki W, Tanaka H, Abe T (2000). Microsatellite analysis of the regeneration process of Magnolia obovata Thunb. Heredity 84: 143–151.
Isagi Y, Suhandono S (1997). PCR primers amplifying microsatellite loci of Quercus myrsinifolia Blume and their conservation between oak species. Mol Ecol 6: 897–899.
Itow S (1991). Species turnover and diversity patterns along an evergreen broad-leaved forest cenocline. J Veg Sci 2: 477–484.
Kalisz S, Nason JD, Hanzawa FM, Tonsor SJ (2001). Spatial population genetic structure in Trillium grandiflorum: the roles of dispersal, mating, history, and selection. Evolution 55: 1560–1568.
Kampfer S, Lexer C, Glossl J, Steinkellner H (1998). Characterization of (GA)n microsatellite loci from Quercus robur. Hereditas 129: 183–186.
Loiselle BA, Sork VL, Nason J, Graham C (1995). Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82: 1420–1425.
Manabe T, Nishimura N, Miura M, Yamamoto S (2000). Population structure and spatial patterns for trees in a temperate old-growth evergreen broad-leaved forest in Japan. Plant Ecol 151: 181–197.
Meagher TR, Thompson E (1986). The relationship between single parent and parent pair genetic likelihoods in genealogy reconstruction. Theor Popul Biol 29: 87–106.
Nakanishi A, Tomaru N, Yoshimaru H, Kawahara T, Manabe T, Yamamoto S (2004). Patterns of pollen flow and genetic differentiation among pollen pools in Quercus salicina in a warm temperate old-growth evergreen board-leaved forest. Silvae Genet 53: 258–264.
Raymond M, Rousset F (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86: 248–249.
Ritland K (1996). Estimators for pairwise relatedness and individual inbreeding coefficients. Genet Res 67: 175–185.
Rousset F (2002). Inbreeding and relatedness coefficients: what do they measure? Heredity 88: 371–380.
Setsuko S, Ishida K, Ueno S, Tsumura Y, Tomaru N (2007). Population differentiation and gene flow within a metapopulation of a threatened tree, Magnolia stellata (Magnoliaceae). Am J Bot 94: 128–136.
Sezen UU, Chazdon RL, Holsinger KE (2005). Genetic consequences of tropical second-growth forest regeneration. Science 307: 891.
Silvertown JW, Charlesworth D (2001). Introduction to Plant Population Biology, 4th edn. Blackwell Science: Oxford.
Smouse PE, Dyer RJ, Westfall RD, Sork VL (2001). Two-generation analysis of pollen flow across a landscape. I. Male gamete heterogeneity among females. Evolution 55: 260–271.
Steinkellner H, Fluch S, Turetschek E, Lexer C, Streiff R, Kremer A et al. (1997). Identification and characterization of (GA/CT)n—microsatellite loci from Quercus petraea. Plant Mol Biol 33: 1093–1096.
Streiff R, Ducousso A, Lexer C, Steinkellner H, Gloessl J, Kremer A (1999). Pollen dispersal inferred from paternity analysis in a mixed oak stand of Quercus robur L. and Q. petraea (Matt.) Liebl. Mol Ecol 8: 831–841.
Valbuena-Carabana M, Gonzalez-Martinez SC, Sork VL, Collada C, Soto A, Goicoechea PG (2005). Gene flow and hybridisation in a mixed oak forest (Quercus pyrenaica Willd. and Quercus petraea (Matts.) Liebl.) in central Spain. Heredity 95: 457–465.
Acknowledgements
Field and laboratory assistance was provided by M Miura, T Fujita and other members of the Laboratory of Forest Ecology and Physiology of Nagoya University whose help is gratefully acknowledged. We thank T Torimaru for his statistical assistance, and the Tsushima District Forest Office for permitting this study, which was supported by Grants-in-Aid for Scientific Research (nos. 11460069 and 14206017) from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nakanishi, A., Tomaru, N., Yoshimaru, H. et al. Effects of seed- and pollen-mediated gene dispersal on genetic structure among Quercus salicina saplings. Heredity 102, 182–189 (2009). https://doi.org/10.1038/hdy.2008.101
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2008.101
Keywords
This article is cited by
-
Genetic diversity in North American Cercis Canadensis reveals an ancient population bottleneck that originated after the last glacial maximum
Scientific Reports (2021)
-
Small but not isolated: a population genetic survey of the tropical tree Cariniana estrellensis (Lecythidaceae) in a highly fragmented habitat
Heredity (2016)
-
Inbreeding depression at the sapling stage and its genetic consequences in a population of the outcrossing dominant tree species, Castanopsis sieboldii
Tree Genetics & Genomes (2015)
-
Genetic evidence of reproductive isolation in a remote enclave of Quercus pubescens in the presence of cross-fertile species
Plant Systematics and Evolution (2012)
-
Low levels of realized seed and pollen gene flow and strong spatial genetic structure in a small, isolated and fragmented population of the tropical tree Copaifera langsdorffii Desf
Heredity (2011)