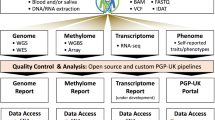
Abstract
The completion of the Human Genome Project (HGP) in 2001 opened the floodgates to a deeper understanding of medicine. There are dozens of HGP-like projects which involve from a few tens to several million genomes currently in progress, which vary from having specialized goals or a more general approach. However, data generation, storage, management and analysis in public and private cloud computing platforms have raised concerns about privacy and security. The knowledge gained from further research has changed the field of genomics and is now slowly permeating into clinical medicine. The new precision (personalized) medicine, where genome sequencing and data analysis are essential components, allows tailored diagnosis and treatment according to the information from the patient’s own genome and specific environmental factors. P4 (predictive, preventive, personalized and participatory) medicine is introducing new concepts, challenges and opportunities. This review summarizes current sequencing technologies, concentrates on ongoing human genomics projects, and provides some examples in which precision medicine has already demonstrated clinical impact in diagnosis and/or treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
NHSEngland. Improving outcomes through personalised medicine. NHS England, 2016. Available from: https://www.england.nhs.uk/wp-content/uploads/2016/09/improving-outcomes-personalised-medicine.pdf.
Goodwin S, McPherson JD, McCombie WR . Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 2016; 17: 333–351.
Illumina. Understanding the genetic code. NGS technology enables massively parallel DNA analysis for a deeper understanding of biology. Illumina, 2017. Available from: https://www.illumina.com/techniques/sequencing/dna-sequencing.html.
Hayden EC . Is the $1000 genome for real?. Nature Publishing Group, 2014, Available from: http://www.nature.com/news/is-the-1-000-genome-for-real-1.14530.
Watson M . HiSeq move over, here comes Nova! A first look at Illumina NovaSeq. Watson M, 2017. Available from: http://www.opiniomics.org/hiseq-move-over-here-comes-nova-a-first-look-at-illumina-novaseq/.
ThermoFisherScientific SOLiD Next-Generation Sequencing. Thermo Fisher Scientific, 2017, Available from: https://www.thermofisher.com/us/en/home/life-science/sequencing/next-generation-sequencing/solid-next-generation-sequencing.html.
ThermoFisherScientific Ion Torrent Next-Generation Sequencing Technology. Thermo Fisher Scientific, 2017, Available from: https://www.thermofisher.com/es/es/home/life-science/sequencing/next-generation-sequencing/ion-torrent-next-generation-sequencing-technology.html.
Qiagen. GeneReader Platform. For next-generation sequencing applications Qiagen 2017, Available from: https://www.qiagen.com/es/products/ngs/mdx-ngs-genereader/library-preparation/qiagen-genereader-platform/.
BGI BGISEQ-500: a BGI Sequencer. Beijing Genomics Institute (BGI), 2017, Available from: http://www.genomics.cn/en/navigation/show_navigation?nid=4203.
PacBioSMRT science. SMRT sequencing Advance genomics with single-molecule, real-time (SMRT) sequencing. Pacific Biosciences (PacBio), 2017, Available from: http://www.pacb.com/smrt-science/smrt-sequencing/.
Robinson K . AGBT2017, PacBio Workshop. Robinson K, 2017. Available from: https://storify.com/OmicsOmicsBlog/agbt2017-wed-pacbio-workshop.
OxfordNanoporeTechnologies. How does nanopore DNA/RNA sequencing work? Oxford Nanopore Technologies, 2017. Available from: https://nanoporetech.com/how-it-works.
Robinson K . ONT Updates: GridION X5, PromethION, 1D^2, Scrappie, FPGAs and More. Robinson K, 2017. Available from: http://omicsomics.blogspot.com.es/2017/03/ont-updates-gridion-x5-promethion-1d2.html#more.
Illumina Long-Read Sequencing Technology. Illumina, 2017, Available from: https://www.illumina.com/science/technology/next-generation-sequencing/long-read-sequencing.html.
Genomics X. The GemCode Technology. A fully integrated solution for RNA and DNA analysis. 10X Genomics, 2017. Available from: https://www.10xgenomics.com/technology/.
iGenomXiGenomX library construction products are purpose built for each applicationThe Ideal Data Set for an Application is Envisioned, then Molecular Methods are Developed to Achieve the Ideal. iGenomX, 2017, Available from: http://www.igenomx.com/technology.html.
SEQLLtSMS Overview. True Single Molecule SequencingWith tSMS, Read What is Really in Sample. SEQLL, 2017. Available from: http://seqll.com/technology-information/.
Genapsys Changing the World, One Genius at a Time. GenapSys, 2017. Available from: http://www.genapsys.com/product2/productwithfeatures.html.
Genia Biological Nanopores: Structure-Based Sequencing of Single DNA Molecules. Genia, 2017. Available from: http://www.geniachip.com/technology/.
Robinson K . Illumina's Unveils Firefly. Robinson K 2016. Available from: http://omicsomics.blogspot.com.es/2016/01/illuminas-unveils-firefly.html.
Yuzuki D . NanoString’s Hyb&Seq single-molecule sequencing platform at AGBT 2017. Yuzuki D 2017. Available from: http://www.yuzuki.org/nanostrings-hyb-seq-single-molecule-sequencing-platform-agbt-2017/#tc-comment-title.
Gnubio The GnuBIO Platform is the only Fully Integrated Next-generation DNA Sequencing Platform. Gnubio, 2017. Available from: http://gnubio.com/technology/.
Mankos M, Shadman K, Persson HH, N'Diaye AT, Schmid AK, Davis RW . A novel low energy electron microscope for DNA sequencing and surface analysis. Ultramicroscopy 2014; 145: 36–49.
Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ et al. Big Data: Astronomical or Genomical? PLoS Biol 2015; 13: e1002195.
Tang H, Jiang X, Wang X, Wang S, Sofia H, Fox D et al. Protecting genomic data analytics in the cloud: state of the art and opportunities. BMC Med Genomics 2016; 9: 63.
Homer N, Szelinger S, Redman M, Duggan D, Tembe W, Muehling J et al. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet 2008; 4: e1000167.
Muddyman D, Smee C, Griffin H, Kaye J . Implementing a successful data-management framework: the UK10K managed access model. Genome Med 2013; 5: 100.
Lappalainen I, Almeida-King J, Kumanduri V, Senf A, Spalding JD, Ur-Rehman S et al. The European Genome-phenome Archive of human data consented for biomedical research. Nat Genet 2015; 47: 692–695.
NCBI. SRA Handbook: National Center for Biotechnology Information (US) 2009. Available from: https://www.ncbi.nlm.nih.gov/books/NBK47533/.
Mashima J, Kodama Y, Kosuge T, Fujisawa T, Katayama T, Nagasaki H et al. DNA data bank of Japan (DDBJ) progress report. Nucleic Acids Res 2016; 44: D51–D57.
Google. https://cloud.google.com/genomics/ Google 2017. Available from: GOOGLE GENOMICS. Ask bigger questions by efficiently processing up to petabytes of genomic data.
Microsoft. Microsoft Genomics. Microsoft 2017. Available from: https://enterprise.microsoft.com/en-us/industries/health/genomics/.
Amazon Genomics in the Cloud. Innovate and Collaborate with Colleagues Around the World by Deploying in the Amazon Web Services Cloud. Amazon, 2017. Available from: https://aws.amazon.com/health/genomics/?nc1=h_ls.
US. DepartmentHealth&HumanServices. Health Information Privacy. U.S. Department of Health & Human Services 2017, Available from: https://www.hhs.gov/hipaa/.
NHGRI. International Human Genome Sequencing Consortium Describes finished human genome sequence Researchers Trim Count of Human Genes to 20,000-25,000. National Human Genome Research Institute, 2004, Available from: https://www.genome.gov/12513430/2004-release-ihgsc-describes-finished-human-sequence/.
Moraes F, Goes A . A decade of human genome project conclusion: Scientific diffusion about our genome knowledge. Biochem Mol Biol Educ 2016; 44: 215–223.
Boeke JD, Church G, Hessel A, Kelley NJ, Arkin A, Cai Y et al. Genome Engineering. The Genome Project-Write. Science 2016; 353: 126–127.
Muddyman D . The UK10K Project: 10000 UK Genome Sequences-Accessing the Role of Rare Genetic Variants in Health and Disease. In: E Zeggini AM ed. Assessing Rare Variation in Complex Traits, 2015, pp 87–105.
WTSI UK10K Rare Genetic Variants in Health and Disease. Wellcome Trust Sanger Institute, 2014, Available from: http://www.uk10k.org/.
Kaye J, Hurles M, Griffin H, Grewal J, Bobrow M, Timpson N et al. Managing clinically significant findings in research: the UK10K example. Eur J Hum Genet 2014; 22: 1100–1104.
GenomicsEngland The 100000 Genomes Project. Genomics England, 2017, Available from: https://www.genomicsengland.co.uk/the-100000-genomes-project/.
GenomeAsia100K GenomeAsia 100K. We are a Mission Driven Non-Profit Consortium Collaborating to Sequence and Analyze 100000 Asian Individuals Genomes to Help Accelerate Asian Population Specific Medical Advances and Precision Medicine. GenomeAsia 100K, 2017, Available from: http://www.genomeasia100k.com/.
Collins FS, Varmus H . A new initiative on precision medicine. N Engl J Med 2015; 372: 793–795.
TheWhiteHouse The Precision Medicine Initiative. The White House President Barack Obama 2015, Available from: https://obamawhitehouse.archives.gov/precision-medicine.
China.org.cn. China kicks off precision medicine research. China.org.cn 2016, Available from: http://www.china.org.cn/china/Off_the_Wire/2016-01/09/content_37537726.htm.
Liu P . China initiative would pour billions into precision medicine. BioWorld. The Daily Biopharmaceutical News Source Now From Thomson Reuters 2016, Available from: http://www.bioworld.com/content/china-initiative-would-pour-billions-precision-medicine-0.
WHO Cancer. World Health Organization, 2017, Available from: http://www.who.int/mediacentre/factsheets/fs297/en/.
ICGC ICGC Cancer Genome Projects. International Cancer Genome Consortium (ICGC), 2016, Available from: https://icgc.org/.
Puente XS, Bea S, Valdes-Mas R, Villamor N, Gutierrez-Abril J, Martin-Subero JI et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 2015; 526: 519–524.
Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016; 534: 47–54.
NCI-NHGRI The Cancer Genome Atlas Program Overview. National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI), 2017, Available from: https://cancergenome.nih.gov/abouttcga/overview.
Cancer Genome Atlas Research N. Integrated genomic and molecular characterization of cervical cancer. Nature 2017; 543: 378–384.
Girotti MR, Gremel G, Lee R, Galvani E, Rothwell D, Viros A et al. Application of Sequencing, Liquid Biopsies, and Patient-Derived Xenografts for Personalized Medicine in Melanoma. Cancer Discov 2016; 6: 286–299.
Vaque JP, Martinez N, Batlle-Lopez A, Perez C, Montes-Moreno S, Sanchez-Beato M et al. B-cell lymphoma mutations: improving diagnostics and enabling targeted therapies. Haematologica 2014; 99: 222–231.
Chan BA, Hughes BG . Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Trans Lung Cancer Res 2015; 4: 36–54.
Orphanet About Rare Diseases. Orphanet, 2012, Available from: http://www.orpha.net/consor/cgi-bin/Education_AboutRareDiseases.php?lng=EN.
GlobalGenes RARE List. Global Genes, 2012, Available from: https://globalgenes.org/rarelist/.
IRDiRC About Us. IRDiRC, 2012, Available from: http://www.irdirc.org/about-us/.
(IRDiRC) IRDRC Current Results of Research. IRDiRC, 2016, Available from: http://www.irdirc.org/rare-diseases-research/current-results-of-research/.
NIH NIH Genome Sequencing Program Targets the Genomic Bases of Common, Rare Disease. National Institutes of Health, 2016, Available from: https://www.nih.gov/news-events/news-releases/nih-genome-sequencing-program-targets-genomic-bases-common-rare-disease.
Chong JX, Buckingham KJ, Jhangiani SN, Boehm C, Sobreira N, Smith JD et al. The Genetic basis of mendelian phenotypes: discoveries, challenges, and opportunities. Am J Hum Genet 2015; 97: 199–215.
AMED Initiative on Rare and Undiagnosed Diseases (IRUD). Japan Agency for Medical Research and Development (AMED), 2017, Available from: http://www.amed.go.jp/en/program/IRUD/.
Beaulieu CL, Majewski J, Schwartzentruber J, Samuels ME, Fernandez BA, Bernier FP et al. FORGE Canada Consortium: outcomes of a 2-year national rare-disease gene-discovery project. Am J Hum Genet 2014; 94: 809–817.
RareDiseaseIndiaAlliance GUaRDIAN Genomics for Understanding Rare Diseases India Alliance Network. Rare Disease India Alliance, 2017, Available from: http://guardian.meragenome.com/home.
ClinGen ClinGen—Clinical Genome Resource. ClinGen, 2017, Available from: https://clinicalgenome.org/about/.
WTSI What is the DDD study?. Wellcome Trust Sanger Institute, 2011, Available from: https://www.ddduk.org/intro.html.
Deciphering Developmental Disorders S. Prevalence and architecture of de novo mutations in developmental disorders. Nature 2017; 542: 433–438.
Global Burden of Disease Study C. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015; 386: 743–800.
AutismSpeaks Genetics and Genomics. Autism Speaks, 2017, Available from: https://www.autismspeaks.org/science/initiatives/autism-genome-project.
De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014; 515: 209–215.
Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 2015; 87: 1215–1233.
Jiang YH, Yuen RK, Jin X, Wang M, Chen N, Wu X et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet 2013; 93: 249–263.
C Yuen RK, Merico D, Bookman M, L Howe J, Thiruvahindrapuram B, Patel RV et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci 2017; 20: 602–611.
WHO Epilepsy. World Health Organization, 2017, Available from: http://www.who.int/mediacentre/factsheets/fs999/en/.
Epi4KConsortium. Epi4K: gene discovery in 4,000 genomes. Epilepsia 2012; 53: 1457–1467.
EpilepsyPhenome/GenomeProject About the Epilepsy Phenome/Genome Project. Epilepsy Phenome/Genome Project, 2006, Available from: http://www.epgp.org/.
Epi4K C, Epilepsy Phenome/Genome P Allen AS Berkovic SF Cossette P Delanty N et al. De novo mutations in epileptic encephalopathies. Nature 2013; 501: 217–221.
Euro Epinomics RES Consortium, Epilepsy Phenome/Genome Project, Epi4K C. De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am J Hum Genet 2014; 95: 360–370.
Freyer C . Year 1 of the Epi25 Collaborative—the first 6,000 epilepsy exomes. Genetics Commission of the International League Against Epilepsy (ILAE), 2017, Available from: http://epilepsygenetics.net/2017/01/10/year-1-of-the-epi25-collaborative-the-first-6000-epilepsy-exomes/.
ADI World Alzheimer Report 2015. Alzheimer’s Disease International (ADI), 2015, Available from: https://www.alz.co.uk/research/WorldAlzheimerReport2015.pdf.
U.Pennsylvania The Alzheimer's Disease Sequencing Project (ADSP). University of Pennsylvania Perelman School of Medicine, 2013, Available from: https://www.niagads.org/adsp/content/home.
CureAlzheimer'sFund Converging on a cure. 9 Areas of Focus Whole Genome Sequencing and Epigenetics. Cure Alzheimer's Fund, 2017, Available from: http://curealz.org/focus/whole-genome-sequencing-and-epigenetics.
US.NLM What is the difference between precision medicine and personalized medicine? What about pharmacogenomics?. US National Library of Medicine, 2017, Available from: https://ghr.nlm.nih.gov/primer/precisionmedicine/precisionvspersonalized.
Schork NJ . Time for one-person trials. Nature 2015; 520: 609–611.
PersonalizedMedicineCoalition Personalized Medicine Coalition. Personalized Medicine Coalition, 2016, Available from: http://www.personalizedmedicinecoalition.org/.
CentreforPersonalisedMedicine Personalised Medicine. Wellcome Trust Centre for Human Genetics, 2017, Available from: http://www.well.ox.ac.uk/cpm/personalised-medicine-2.
Kienzler AK, Hargreaves CE, Patel SY . The role of genomics in common variable immunodeficiency disorders. Clin Exp Immunol 2017; 188: 326–332.
Cookson C Genomics promises a leap forward for rare disease diagnosis Faster and Cheaper DNA Sequencing Brings New Hope to Patients. Finantial Times, 2017, Available from: https://www.ft.com/content/d2e21cea-d684-11e6-944b-e7eb37a6aa8e.
UDN The Undiagnosed Diseases Network (UDN). The Undiagnosed Diseases Network (UDN) with support from the NIH Common Fund, 2016, Available from: https://undiagnosed.hms.harvard.edu/.
Ramoni RB, Mulvihill JJ, Adams DR, Allard P, Ashley EA, Bernstein JA et al. The undiagnosed diseases network: accelerating discovery about health and disease. Am J Hum Genet 2017; 100: 185–192.
Heger M . Despite Actionable Results, Cancer Sequencing Projects Struggle to Get Patients on Targeted Drugs. GenomeWeb LLC, 2017, Available from: https://www.genomeweb.com/cancer/despite-actionable-results-cancer-sequencing-projects-struggle-get-patients-targeted-drugs.
McCarthy MI . Painting a new picture of personalised medicine for diabetes. Diabetologia 2017; 60: 793–799.
De Bellis M, Conte Camerino D, Desaphy JF . Toward precision medicine in myotonic syndromes. Oncotarget 2017; 8: 14279–14280.
Sagner M, McNeil A, Puska P, Auffray C, Price ND, Hood L et al. The P4 Health Spectrum—A Predictive, Preventive, Personalized and Participatory Continuum for Promoting Healthspan. Prog Cardiovasc Dis 2016; 59: 506–521.
Rabbani B, Nakaoka H, Akhondzadeh S, Tekin M, Mahdieh N . Next generation sequencing: implications in personalized medicine and pharmacogenomics. Mol Biosyst 2016; 12: 1818–1830.
Salvi S, Gurioli G, De Giorgi U, Conteduca V, Tedaldi G, Calistri D et al. Cell-free DNA as a diagnostic marker for cancer: current insights. Onco Targets Ther 2016; 9: 6549–6559.
Volik S, Alcaide M, Morin RD, Collins C . Cell-free DNA (cfDNA): clinical significance and utility in cancer shaped by emerging technologies. Mol Cancer Res 2016; 14: 898–908.
Hood L, Auffray C . Participatory medicine: a driving force for revolutionizing healthcare. Genome Med 2013; 5: 110.
InstituteforSystemsBiology P4 Medicine. Institute for Systems Biology, 2017, Available from: https://www.systemsbiology.org/research/p4-medicine/.
Murray TH, Livny E . The Human Genome Project: ethical and social implications. Bull Med Libr Assoc 1995; 83: 14–21.
Park ST, Kim J . Trends in next-generation sequencing and a new era for whole genome sequencing. Int Neurourol J 2016; 20 (Suppl 2): S76–S83.
MedicineGov Global Alliance for Genomics and Health. Medicine Gov, 2017, Available from: http://medicinegov.org/global-alliance-for-genomics-and-health/#sthash.q0pfnXVI.CtkPnL4J.dpbs.
Acknowledgements
We thank our Genomics and Next Generation Sequencing Service colleagues for their invaluable professional work, expertize and discussions. Without them this review could not have been written. We thank all the researchers (our ‘customers’) who place their trust on us and keep our motivation to continuously acquire new knowledge in this fascinating field. We also thank the reviewers for their constructive suggestions. We want to apologize to all those whom we have not cited in this review due to space constraints. The CBMSO receives institutional grants from the Fundación Ramón Areces and from the Fundación Banco de Santander.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Carrasco-Ramiro, F., Peiró-Pastor, R. & Aguado, B. Human genomics projects and precision medicine. Gene Ther 24, 551–561 (2017). https://doi.org/10.1038/gt.2017.77
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gt.2017.77
This article is cited by
-
Understanding genetic variability: exploring large-scale copy number variants through non-invasive prenatal testing in European populations
BMC Genomics (2024)
-
Antioxidant evaluation and computational prediction of prospective drug-like compounds from polyphenolic-rich extract of Hibiscus cannabinus L. seed as antidiabetic and neuroprotective targets: assessment through in vitro and in silico studies
BMC Complementary Medicine and Therapies (2023)
-
A display and analysis platform for gut microbiomes of minority people and phenotypic data in China
Scientific Reports (2023)
-
Identification of novel missense mutation related with non-syndromic sensorineural deafness, DFNA11 in korean family by NGS
Genes & Genomics (2023)
-
Understanding Individual Subject Differences through Large Behavioral Datasets: Analytical and Statistical Considerations
Perspectives on Behavior Science (2023)