Abstract
Purpose:
The aim of this study was to survey American College of Medical Genetics and Genomics members about secondary findings from clinical genome-scale sequencing.
Methods:
A Web-based survey was mailed to 1,687 members of the American College of Medical Genetics and Genomics. Exploratory factor analysis identified underlying factors assessed by survey items. Linear regression assessed associations between factor scores and respondent characteristics.
Results:
The response rate was 29%. Four factors explained 51% of the survey variance: best practices, patient preferences, guidance, and informed consent. Most agreed with “best practice” items describing seeking and reporting of secondary findings as consistent with medical standards, having sufficient evidence, and, for adults, the benefits generally outweighing potential harms. There was lack of agreement regarding benefits versus harms for children and impact on health-care resources. The majority agreed that patient preferences should be considered, including ability to opt out, and that informed consent was feasible and critical. Characteristics significantly associated with factor scores included country of residence, sequencing experience, and years in practice.
Conclusion:
The American College of Medical Genetics and Genomics should update a list of genes to be assessed when clinical genome-scale sequencing is performed. Informed consent is necessary, and reporting of secondary findings should be optional. Research on implementation of secondary findings reporting is needed.
Genet Med advance online publication 13 November 2014
Similar content being viewed by others
Introduction
Genome-scale analysis is rapidly being incorporated into clinical genetic practice.1,2 According to a recent American College of Medical Genetics and Genomics (ACMG) policy statement, exome or genome sequencing should be considered for diagnostic purposes when a genetic etiology is likely but targeted testing is not available; a patient’s disorder has high genetic heterogeneity; targeted testing resulted in no diagnosis; or a fetus has a disorder that is likely genetic but remains undiagnosed.3
Next-generation sequencing offers the advantages of speed and efficiency relative to Sanger sequencing of individual genes. It can also characterize new diseases or genetic syndromes by identifying pathogenic variants in novel or candidate genes and further define phenotypic aspects of genetic disorders.4,5,6 However, as is true with Sanger (or single-gene) sequencing, massively parallel sequencing has limitations; certain pathogenic variants are missed and variants of uncertain significance often are detected.7 As with other broad genetic tests, when sequencing the exome or genome, findings unrelated to the original indication for testing are identified. Occasionally, pathogenic variants are identifiable that could alert patients and providers to important medical conditions not previously suspected in the individual. Such findings have been variously labeled as medically actionable “incidental” or “secondary” findings, and precisely how to deal with them in terms of analysis and reporting has been a matter of considerable debate.8,9,10 For the purposes of this article, we follow the recommendation of the Presidential Commission on Bioethical Issues and use the term “secondary finding,” which is defined as a result that is not the primary target of the test but an additional result that is actively sought, as recommended by an expert body or by a consensus of practitioners.11
An ACMG working group was launched in 2012 to evaluate the need for, and the principles that would govern, recommendations for analyzing and reporting secondary findings from exome or genome sequencing in the clinical context and to generate an initial list of genes and categories of variants that should be reported as secondary findings based on the general concept of medical action ability.12 The working group considered a number of issues in their deliberations, including the clinical utility of secondary findings, the limitations of interpreting such findings, patient preferences, and special considerations when sequencing children. Moreover, they excluded from their deliberations certain issues, including sequencing for preconception testing, prenatal testing, and newborn screening, and sequencing of healthy children and adults.
In March 2013 the working group published five recommendations regarding the analysis and reporting of medically actionable secondary findings from clinical exome and genome sequencing. The working group recommended that sequencing laboratories routinely detect and report secondary findings in a minimal list of “medically actionable” genes that are known to cause a disorder, regardless of the indication for sequencing and regardless of the age of the patient, without offering the opportunity for patients to opt out of such analysis. Furthermore, they recommended that this subset of secondary findings be sought and reported for any normal sequenced germline tissue, including normal tissue from a tumor–normal dyad, and for unaffected members of a family trio. These recommendations were an attempt “to strike a balance between the positions of genetic libertarians and the genetic empiricists, guided by the currently available scientific literature, clinical experience, the consensus of [the] Working Group members, and the traditions of clinical medicine.”12 The working group also recommended that the ordering clinician or team provide comprehensive counseling to the patient before and after testing, and that the ACMG, together with content experts and other professional organizations, refine and update annually the minimal list of medically actionable genes and their respective conditions.
Response from the genetics community to these recommendations was mixed. In July 2013 the ACMG Board of Directors agreed that additional information from the ACMG membership was needed to ensure that the working group recommendations would be in keeping with a consensus of membership. A plan to survey the opinions of ACMG’s membership was made, the results of which would inform further ACMG recommendations regarding these issues.
Materials and Methods
Survey development
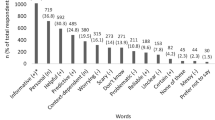
A survey team comprising members of the ACMG Board of Directors and staff with expertise in clinical and molecular genetics and survey development was assembled. The ACMG director of public relations identified 26 information sources published between 21 March 2013 and 24 July 2013 in response to the ACMG recommendations issued on 21 March 2013. Only written documents were included for review; audio interviews and webinars without transcripts were excluded. The survey team reviewed these information sources and concluded that reactions to the ACMG recommendations were mostly grounded in one or more ethical arguments. As a result, one reviewer (M.T.S.) categorized reaction statements according to the four principles of medical ethics13: autonomy, beneficence, nonmaleficence, and justice (including distributive, rights-based, and legal justice). Quotes from published sources were entered into a matrix for rapid thematic analysis. One team member (M.T.S.) identified themes within each ethical construct, which were then reviewed and discussed by other team members until consensus was reached. A draft version of the survey was developed using Research Electronic Data Capture software (http://www.project-redcap.org). Pilot testing, including cognitive and usability testing, was then conducted with nine individuals including two trainees and seven medical geneticists with expertise in clinical genetics (n = 4), molecular genetics (n = 2), cytogenetics (n = 2), and genetic counseling (n = 1) (some had more than one area of expertise). Revisions to the survey resulted in 11 survey items describing respondent characteristics, an open-ended item asking about tools and resources needed, and 33 items addressing the themes identified, each with a five-point scaled response assessing level of agreement (ranging from 1 = strongly disagree to 5 = strongly agree).
Sample and data collection
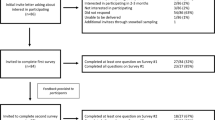
All ACMG members were eligible to complete the survey. An announcement regarding the survey was included in the ACMG e-zine sent to all ACMG members with an e-mail address, and information about the survey was available at the ACMG booth at the American Society of Human Genetics annual meeting in October 2013. In addition, just before launching the survey, an introductory e-mail from the ACMG president was sent to all ACMG members to encourage participation. An e-mail with a survey link was delivered to 1,687 ACMG members on 7 January 2014, with up to five reminder e-mails sent to nonresponders. Data collection closed on 7 March 2014.
Data analysis
Descriptive statistics summarized the responses, exploratory factor analysis identified the most relevant underlying topics (factors) assessed by the items, and linear regression analysis assessed associations between each factor score and respondent characteristics. Negatively worded items were reverse coded.
Principal factor component methods initially yielded a nine-factor solution. Scree plot and parallel factor analysis best supported a six- or four-factor solution. Review of factor loadings resulted in the selection of a four-factor solution. The minimum amount of data for factor analysis was satisfied, with a final sample size of 432 (using listwise deletion), providing a ratio of 15 cases per variable. Survey items with absolute correlation values <0.4 were suppressed; as a result, three items were dropped because of nonloading. In addition, two items that loaded across multiple factors were dropped. The factorability of the 28 remaining items was examined using a correlation matrix and was determined to be suitable based on the following criteria: (i) all items had a correlation of at least 0.3 with at least one other item, suggesting reasonable factorability; (ii) the Kaiser–Meyer–Olkin measure of sampling adequacy was 0.85, much above the commonly recommended value of 0.6; and (iii) Bartlett’s test of sphericity was significant (χ2(378) = 5,757.3; P < 0.05). Two investigators (M.T.S. and J.P.) labeled factors according to the themes that emerged from the items. For each of the four factors, the statistical software program generated a score for each respondent that reflected the aggregate mean score of survey items loading in each factor. Higher scores indicated greater agreement or endorsement. Factor scores were used for linear regression analysis to assess associations between each factor and respondent characteristics. All statistical analyses were performed using STATA statistical software version 13.1 (StataCorp, College Station, TX; http://www.stata.com).
Results
Characteristics of survey respondents
There were 492 respondents (29% response rate). The characteristics of respondents are shown in Table 1 . The gender of the survey respondents (40% men and 56% women) was similar to that of the ACMG membership as of 31 December 2013 (42% men and 58% women). Compared with the ACMG membership, more laboratory geneticists (48 vs. 37%; P < 0.01) and fewer clinical geneticists (38 vs. 45%; P < 0.01) responded to the survey. The frequencies of trainees and students responding to the survey were similar to those of the ACMG membership (7 and 9%, respectively), as were the frequencies of survey respondents from outside the United States and Canada (both 4%). Most of the respondents had read the ACMG recommendations and were familiar with the ACMG minimal list of genes to be analyzed and reported with clinical exome or genome sequencing. In addition, 57% had ordered or performed exome or genome sequencing at least once in the past 12 months.
Factor analysis
Exploratory factor analysis identified four factors that explained 51% of the survey variance. The interpretation of the exploratory factor analysis is shown in Table 2 , including the characterization of each factor based on the survey items identified and the percentage of the variance of survey responses explained by each factor. Survey items included in each factor and frequencies of item responses are shown in Table 3 . For each item, a small percentage of respondents, ranging from 0.6 to 3%, did not respond.
The 16 survey items contributing to factor 1 address “clinical and laboratory best practices,” that is, what should and should not be done and what could and could not happen as a result of seeking and reporting secondary findings. Most respondents agreed that seeking and reporting secondary findings for medically actionable genes is consistent with medical standards and that there is sufficient evidence to report these variants; in addition, most respondents agreed that the benefits of seeking and reporting secondary findings for medically actionable genes generally outweigh potential harms for adults. There was, however, lack of agreement on this issue for children. A small percentage of respondents were neutral to the idea of seeking and reporting secondary findings for adults and children, and a minority thought it should never be done. Responses about the potential impact of seeking and reporting secondary findings on health-care resources were mixed, with 26–33% neutral on the matter; similar percentages of respondents agreed and disagreed that there would be an impact.
Factor 2 consists of six survey items that describe “patient preferences.” The majority agreed that patients should be able to opt out of receiving secondary findings; that not consenting to receiving secondary findings should not influence whether a patient is offered clinical exome or genome sequencing; and that denying exome or genome sequencing if a patient declines to receive secondary findings is coercive. About half believed patients should be able to customize which of the genes on the ACMG list are included in their report, and about half also believed that a laboratory should customize the report according to patient preferences regarding the genes on the ACMG list. Only 7–17% of respondents were neutral regarding these items.
Three survey items included in factor 3 relate to “guidance for practice”; these items describe the benefit of maintaining a list of genes that should be included when reporting secondary findings when exome or genome sequencing is performed. Few respondents were neutral on these items, and most agreed it would be beneficial to have and update a minimal list of genes for which medically actionable secondary findings should be sought and reported if experts deem the associated conditions medically actionable.
Factor 4 includes three survey items that address “informed consent.” A large majority believed the ordering clinician is responsible for pre- and posttest counseling and education regarding secondary findings that could be identified in one or more of the genes on the ACMG minimal list and that informed consent to seek and report such secondary findings is feasible (75%) and critical (79%); only 9–12% of respondents were neutral on these items.
Regression analysis
There were no significant associations between any of the factor scores and respondent characteristics of sex, specialty, trainee status, or percentage of time spent in direct patient care. There were also no significant differences between respondents who had read the ACMG recommendations or were familiar with the ACMG minimal list of genes for secondary findings and those who had not/were not, but few had not read the report or were not familiar with the minimal list of genes.
Country of residence was the only characteristic significantly associated with level of agreement regarding best practice items (factor 1). Compared with respondents living or working in the United States, those from a different country were significantly less likely to agree with best practice items (b = −0.51; t(388) = −2.51; P = 0.012).
The most significant associations with respondent characteristics were observed for items addressing patient preferences (factor 2). Experience with ordering or performing exome or genome sequencing was significantly and inversely associated with the level of agreement regarding such preferences. Compared with respondents who had never ordered or performed sequencing, respondents who had ordered a few (1–5) or many (>20) such tests in the past year had significantly lower scores supporting patient preferences (b = −0.44; t(388) = −2.93; P = 0.004 and b = −0.43; t(388) = −2.41; P = 0.016, respectively). They were less likely to agree that the patient should be able to opt out of receiving a report with secondary findings, that the patient should have the ability to choose which genes are included in the report of secondary findings, or that the laboratory should customize the reporting of secondary findings based on patient preferences. On the other hand, working at or using a sequencing laboratory that provides an option for receipt of secondary findings was positively associated with patient preferences (b = 0.46; t(388) = 3.24; P = 0.001).
The number of years in practice (a surrogate for the respondent’s age) was the only characteristic significantly associated with level of agreement regarding informed consent items (factor 4). Compared with respondents with no clinical experience, we saw a significant increase in the level of agreement regarding the necessity of informed consent for the possibility of secondary findings among the youngest practitioners (1–10 years of experience; b = 0.60; t(388) = 2.47; P = 0.014), with a statistically nonsignificant decreasing trend in agreement as years of experience increased.
There were no significant associations between respondent characteristics and the level of agreement with survey items related to guidance for clinical practice (factor 3). However, we did observe a statistically nonsignificant trend: as the number of sequencing tests ordered or performed in the past year increased, respondents were less likely to agree with the need for practice guidance.
Discussion
This survey of the ACMG membership corroborates many of the recommendations formulated by the ACMG working group.12 Namely, clinical exome or genome sequencing laboratories should seek and report secondary findings for genes considered to be medically actionable; the ACMG, together with content experts and other professional organizations, should refine and update this list; and clinicians ordering these tests should provide pre- and posttest counseling and education for the possibility of secondary findings. However, contrary to the ACMG recommendations, the majority of respondents believed that patient preferences regarding seeking and reporting of secondary findings should be considered when clinical exome or genome sequencing is pursued, including the ability of patients to opt out of receiving a sequencing report with such findings. A similar survey of genetics professionals also found that, “The vast majority (81%) thought that individual preferences should guide return [of secondary findings].”10
Analysis of survey items that were informed by medical ethical principles revealed four factors or underlying topics of importance to ACMG members, including (i) best practices surrounding clinical exome or genome sequencing; (ii) respect for patient preferences regarding secondary findings when performing exome or genome sequencing; (iii) the need for clinician and laboratory guidance regarding secondary findings; and (iv) the informed consent process for secondary findings. These constructs should be considered as the ACMG and other organizations address issues pertaining to analysis and reporting of secondary findings from exome or genome sequencing. In this study none of the factor scores was significantly associated with respondent specialty, sex, trainee status, or time spent in direct patient care. The lack of association with specialty is particularly important because the ACMG recommendation placed different responsibilities on clinical and laboratory geneticists with respect to seeking and reporting secondary findings from clinical genome-scale sequencing.12
Lack of agreement among survey items identified topics that would benefit from additional investigation and the generation of empirical data to inform practice and policy. The National Human Genome Research Institute of the National Institutes of Health is already funding clinical sequencing exploratory research centers that are conducting multidisciplinary investigations of clinical and health services issues arising from the return of sequencing results in both clinical and research settings.14
Within the “best practice” construct, there was no consensus regarding seeking and reporting of secondary findings when exome or genome sequencing is performed on children or on parents when trio testing is done. Of note, the Presidential Commission on Bioethical Issues did not address the reporting of secondary findings in children.11 In addition, there was no consensus regarding the impact of seeking and reporting secondary findings on health-care resources, including the clinical workforce and access to genetic services. These, unlike many other issues surrounding the recommendations, are essentially empirical questions. ACMG members working outside of the United States were significantly less likely to agree with the best practice items compared with those working in the United States. This may reflect differences in health-care systems, including regulations governing genetic and genomic testing, health professional genetics education and training, and access to genetic health care and other health-care resources.15 Therefore these external and organizational factors should be considered when planning the implementation of reporting secondary findings from exome or genome sequencing.
Concerning the “patient preference” construct, although there was overwhelming agreement that patients should be able to opt out of receiving a report with secondary findings, there was no consensus regarding customization of a secondary findings list of genes according to patient preferences. Interestingly, increasing experience with exome or genome sequencing was inversely associated with deferring to patient preferences. Perhaps those with more experience recognize the nuances and potential difficulties of customizing a list, particularly one that is likely to grow and change over time.
A substantial majority of respondents agreed that the process of informed consent for secondary findings from exome or genome sequencing is critical and feasible. Individuals with fewer years of clinical practice, who are presumably younger, were significantly more likely to agree that informed consent was necessary. The reasons for this finding are likely multiple but may, in part, be a consequence of differences in training—emphasis on informed consent and ethical issues has become more prominent in recent years16—or perhaps clinicians and laboratorians with more years of experience may perceive informed consent for possible secondary findings as impractical. Understanding these differences could be important to successfully developing and implementing informed consent for secondary findings. Tools and templates that assist clinicians in performing and documenting the informed consent process for secondary findings likely would be useful.
The survey response rate was 29%, and in many respects the characteristics of respondents were similar to those of the ACMG membership, supporting good representation of membership views. Furthermore, survey results were consistent with the results of other recent surveys of genetics professionals regarding the reporting of medically actionable secondary findings when exome or genome sequencing is performed.10,17,18 In addition, the response rate was more than adequate to perform the exploratory factor analysis that identified the four major topics revealed by the survey items and that can inform future studies and policy development in this area. Indeed, based on these survey results, the ACMG has updated its position on the issue of opting out of receiving a report containing secondary findings.19
In summary, survey results indicate that the ACMG membership generally believes that the ACMG should continue to update a minimal list of medically actionable genes and conditions that should be considered when clinical exome or genome sequencing is performed. However, reporting of secondary findings should be optional and included in exome or genome sequencing reports only with the consent of the patient or the patient’s guardian. Survey results also identified items for consideration regarding best practices for the laboratory and clinician. Moreover, survey results identified gaps where research is needed to understand multilevel factors, including patient, provider, organizational, and external factors that may influence successful implementation of seeking and reporting secondary findings when clinical exome or genome sequencing is performed. Such gaps include the impact of reporting secondary findings on health and psychosocial outcomes (particularly for children), as well as on health-care outcomes, including utilization of health-care resources and resulting consequences for the health-care workforce and access to genetic services.
Disclosure
The authors declare no conflict of interest.
References
Manolio TA, Chisholm RL, Ozenberger B, et al. Implementing genomic medicine in the clinic: the future is here. Genet Med 2013;15:258–267.
Biesecker LG, Green RC . Diagnostic clinical genome and exome sequencing. N Engl J Med 2014;370:2418–2425.
American College of Medical Genetics and Genomics Board of Directors. Points to consider in the clinical application of genomic sequencing. Genet Med 2012;14:759–761.
Shameer K, Klee EW, Dalenberg AK, Kullo IJ . Whole exome sequencing implicates an INO80D mutation in a syndrome of aortic hypoplasia, premature atherosclerosis, and arterial stiffness. Circ Cardiovasc Genet e-pub ahead of print 13 August 2014.
Chen PC, Yin J, Yu HW, et al. Next-generation sequencing identifies rare variants associated with Noonan syndrome. Proc Natl Acad Sci USA 2014;111:11473–11478.
Gee HY, Ashraf S, Wan X, et al. Mutations in EMP2 cause childhood-onset nephrotic syndrome. Am J Hum Genet 2014;94:884–890.
Kuhlenbäumer G, Hullmann J, Appenzeller S . Novel genomic techniques open new avenues in the analysis of monogenic disorders. Hum Mutat 2011;32:144–151.
Grove ME, Wolpert MN, Cho MK, Lee SS, Ormond KE . Views of genetics health professionals on the return of genomic results. J Genet Couns 2014;23:531–538.
Strong KA, Zusevics KL, Bick D, Veith R . Views of primary care providers regarding the return of genome sequencing incidental findings. Clin Genet 2014;86:461–468.
Yu JH, Harrell TM, Jamal SM, Tabor HK, Bamshad MJ . Attitudes of genetics professionals toward the return of incidental results from exome and whole-genome sequencing. Am J Hum Genet 2014;95:77–84.
Presidential Commission for the Study of Bioethical Issues (PCSBI). Anticipate and Communicate: Ethical Management of Incidental and Secondary Findings in the Clinical, Research, and Direct-to- Consumer Contexts. Washington, DC, 2013:27.
Green RC, Berg JS, Grody WW, et al.; American College of Medical Genetics and Genomics. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013;15:565–574.
Beauchamp TL, Childress JF . Principles of Biomedical Ethics, 6th edn, Oxford University Press: New York, 2008.
National Human Genome Research Institute. Clinical Sequencing Exploratory Research (CSER). http://www.genome.gov/CSER. Accessed 15 August 2014.
Battista RN, Blancquaert I, Laberge AM, van Schendel N, Leduc N . Genetics in health care: an overview of current and emerging models. Public Health Genomics 2012;15:34–45.
National Bioethical Advisory Commission. Ethical and Policy Issues in Research Involving Human Participants, summary. Bethesda, MD, 2001. ISBN 1-931022-18-6.
Lohn Z, Adam S, Birch P, Townsend A, Friedman J . Genetics professionals’ perspectives on reporting incidental findings from clinical genome-wide sequencing. Am J Med Genet A 2013;161A:542–549.
Lemke AA, Bick D, Dimmock D, Simpson P, Veith R . Perspectives of clinical genetics professionals toward genome sequencing and incidental findings: a survey study. Clin Genet 2013;84:230–236.
American College of Medical Genetics and Genomics. ACMG News. ACMG Updates Recommendation on “Opt Out” for Genome Sequencing Return of Results. https://www.acmg.net/docs/Release_ACMGUpdatesRecommendations_final.pdf. Accessed 15 August 2014.
Acknowledgements
The authors thank Christopher Day, Megan Lyon, and Meredith Weaver for their input on survey design, development, and data collection. Each provided these services in their role as employees at the ACMG. The contents of this publication are solely the responsibility of the authors and do not represent the official views of the Veterans Health Administration.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Scheuner, M., Peredo, J., Benkendorf, J. et al. Reporting genomic secondary findings: ACMG members weigh in. Genet Med 17, 27–35 (2015). https://doi.org/10.1038/gim.2014.165
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2014.165
Keywords
This article is cited by
-
Ethical values supporting the disclosure of incidental and secondary findings in clinical genomic testing: a qualitative study
BMC Medical Ethics (2020)
-
A proposal on the first Japanese practical guidance for the return of individual genomic results in research settings
Journal of Human Genetics (2020)
-
A pediatric perspective on genomics and prevention in the twenty-first century
Pediatric Research (2020)
-
Incidental or secondary findings: an integrative and patient-inclusive approach to the current debate
European Journal of Human Genetics (2018)
-
Effect of Public Deliberation on Attitudes toward Return of Secondary Results in Genomic Sequencing
Journal of Genetic Counseling (2017)