Abstract
Spinal muscular atrophy is a common autosomal recessive neuromuscular disorder caused by mutations in the survival motor neuron (SMN1) gene, affecting approximately 1 in 10,000 live births. The disease is characterized by progressive symmetrical muscle weakness resulting from the degeneration and loss of anterior horn cells in the spinal cord and brainstem nuclei. The disease is classified on the basis of age of onset and clinical course. Two almost identical SMN genes are present on 5q13: the SMN1 gene, which is the spinal muscular atrophy-determining gene, and the SMN2 gene. The homozygous absence of the SMN1 exon 7 has been observed in the majority of patients and is being used as a reliable and sensitive spinal muscular atrophy diagnostic test. Although SMN2 produces less full-length transcript than SMN1, the number of SMN2 copies has been shown to modulate the clinical phenotype. Carrier detection relies on the accurate determination of the SMN1 gene copies. This document follows the outline format of the general Standards and Guidelines for Clinical Laboratories. It is designed to be a checklist for genetic testing professionals who are already familiar with the disease and methods of analysis.
Similar content being viewed by others
Main
Disease-specific statements are intended to augment the current general American College of Medical Genetics (ACMG) Standards and Guidelines for Clinical Genetic Laboratories. Individual laboratories are responsible for meeting the CLIA/College of American Pathologists (CAP) quality assurance standards with respect to appropriate sample documentation, assay validation, general proficiency, and quality control measures.
BACKGROUND ON SPINAL MUSCULAR ATROPHY
Gene symbol/chromosome locus
Survival motor neuron 1 (SMN1) gene at chromosome 5q11.2-13.3.
OMIM #
Spinal muscular atrophy (SMA) type I (253300), SMA type II (253550), and SMA type III (253400).
Brief clinical description
The autosomal recessive disorder proximal SMA is a severe neuromuscular disease characterized by degeneration of alpha motor neurons in the spinal cord, which results in progressive muscle weakness and paralysis. The predominant pathologic feature on autopsy studies of patients with SMA is loss of motor neurons in the ventral horn of the spinal cord and in brainstem motor nuclei. SMA is the second most common fatal autosomal recessive disorder after cystic fibrosis, with an estimated incidence of approximately 1 in 10,000 live births.1 Childhood SMA is subdivided into three clinical groups on the basis of age of onset and clinical course.2,3 Type I SMA (Werdnig-Hoffman disease) is characterized by severe, generalized muscle weakness and hypotonia at birth or within the first 3 months. Death from respiratory failure usually occurs within the first 2 years. Approximately 60–70% of patients with SMA have the type I disease.4 Type II children are able to sit, although they cannot stand or walk unaided, and typically survive beyond 4 years. The phenotypic variability exceeds that observed in type I patients, ranging from infants who sit transiently and demonstrate severe respiratory insufficiency to children who can sit, crawl, and even stand with support. Prognosis in this group is largely dependent on the degree of respiratory involvement. Type III SMA (Kugelberg-Welander disease) is a milder form, with onset during infancy or youth: patients learn to walk unaided and have prolonged survivals. They comprise a less fragile group than type II patients with respect to respiratory and nutritional vulnerability. Type III SMA is further subdivided into two groups: type IIIa (onset before 3 years of age) and type IIIb (onset at age ≥3 years). Cases presenting with the first symptoms of the disease at the age of 20–30 years are classified as type IV or proximal adult type SMA. The described classification is based on age of onset and clinical course, but it should be recognized that the disorder demonstrates a continuous range of severity. For more information, see the online Gene Reviews profile at http://www.ncbi.nlm.nih.gov/books/NBK1352/.
Mode of inheritance
Inheritance is autosomal recessive, with variable expression.
Gene description/normal gene product
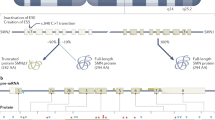
The SMA gene is within a complex region containing multiple repetitive and inverted sequences.5 The SMN gene (Entrez Gene ID number 6606) comprises nine exons with a stop codon present near the end of exon 7.6 Two inverted SMN copies are present: the telomeric or SMN1 gene, which is the SMA-determining gene and the centromeric or SMN2 gene. The two SMN genes are highly homologous, have equivalent promoters, and only differ at five base pairs.5,7,8 The base differences are used to differentiate SMN1 from SMN2. The coding sequence of SMN2 differs from that of SMN1 by a single nucleotide (840C>T), which does not alter the amino acid but has been shown to be important in splicing. SMA results from a reduction in the amount of the SMN protein, and there is a strong correlation between the disease severity and SMN protein levels.9,10 The SMN protein is a ubiquitously expressed, highly conserved 294-amino acid polypeptide. The protein is found in both the cytoplasm and nucleus and is concentrated in punctate structures called “gems” in the nucleus. High levels of the protein have been found to exist in the spinal motor neurons, the affected cells in patients with SMA. The protein self-associates into multimeric structures. Biochemically, SMN does not seem to exist within cells in isolation but instead forms part of a large protein complex, the SMN complex. Many of these SMN interacting proteins are components of various ribonuclear protein (RNP) complexes that are involved in distinct aspects of RNA metabolism. The best characterized function of the SMN complex is regulating the assembly of a specific class of RNA-protein complexes, the small nuclear RNPs (snRNPs).11,12 The snRNPs are a critical component of the spliceosome; a large RNA protein that catalyzes premRNA splicing. SMA may, therefore, be a disorder resulting from aberrant splicing. As the SMN protein is ubiquitously expressed, it remains unknown how a loss of a general housekeeping function (snRNP assembly) causes a selective loss of motor neurons in SMA.13 The high expression of SMN protein in motor neurons may suggest that the neuronal population is more sensitive to decreases in the SMN protein level. Possibly, the altered splicing of a unique set of premessenger RNAs results in deficient proteins, which are necessary for motor neuron growth and survival. In addition to its role in spliceosomal ribonucleoprotein assembly, SMN may have other functions in motor neurons. A subset of SMN complexes is located in axons and growth cones of motor neurons and may be involved in some aspects of axonal transport and localized translation of specific messenger RNAs.14,15
The SMN1 mutation
Homozygous mutations of the SMN1 gene cause SMA. Both copies of the SMN1 gene are absent in approximately 95% of affected patients, whereas the remaining patients have nonsense, frameshift, or missense mutations within the gene.16 Based on Hardy-Weinberg equilibrium, the remaining patients (with the smaller types of mutations) are virtually all assumed to be hemizygous for the SMN1 deletion. The absence of SMN1 can occur by deletion, typically a large deletion that includes the whole gene or by conversion to SMN2. Although patients with SMA have mutations in SMN1, they always carry at least one normal copy of SMN2, which is partially functional but unable to fully compensate for the deficiency of the SMN1 protein. The homozygous loss of both genes has not been reported, presumably as a result of lethality.
Genotype/phenotype association
SMN1 exon 7 is absent in the majority of patients independent of the severity of SMA. Several studies have shown that the SMN2 copy number modifies the severity of the disease.17–20 The SMN2 copy number varies from 0 to 3 copies in the normal population, with approximately 10–15% of normal individuals having no SMN2. However, patients with a milder phenotype with type II or III SMA have been shown to often have more copies of SMN2 than type I patients. The majority of patients with the severe type I form have one or two copies of SMN2; most patients with type II have three SMN2 copies; and most patients with type III have three or four SMN2 copies. Three unaffected family members of patients with SMA, with confirmed SMN1 homozygous deletions, were shown to have five copies of SMN2.21 These cases not only support the role of SMN2 modifying the phenotype but they also demonstrate that expression levels consistent with five copies of the SMN2 genes may be sufficient to compensate for the absence of the SMN1 gene. This inverse dose-relationship between SMN2 copy number and disease severity has also been supported by the SMA mouse model.22,23 The SMA mouse models have not only confirmed the susceptibility of motor neuron degeneration to SMN deficiency but have also verified that the degeneration can be prevented by increased SMN2 dosage. Mice lacking the endogenous mouse SMN gene but expressing two copies of the human SMN2 gene develop severe SMA and die within 1 week of age; however, mice that express multiple copies of SMN2 do not develop the disease. In addition to the SMN2 copy number, other modifying factors influence the phenotypic variability of SMA. There are very rare families reported in which markedly different degrees of disease severity are present in affected siblings with the same SMN2 copy number. These discordant sib pairs, which share the same genetic background around the SMA locus, would indicate that there are other modifier genes outside the SMA region. Differences in splicing factors may allow more full-length expression from the SMN2 gene and account for some of the variability observed between discordant sibs.24 It was also found that in some rare families with unaffected SMN1-deleted females, the expression of plastin 3 was higher than in their SMA affected counterparts.25 Plastin 3 was shown to be important for axonogenesis and, therefore, may act as a protective modifier.
Mutational mechanism
The SMN1 gene produces full-length transcript, whereas the SMN2 gene produces predominantly an alternatively spliced transcript (exon 7 deleted) encoding a protein (SMNΔ7) that does not oligomerize efficiently and is unstable.26,27 The inclusion of exon 7 in SMN1 transcripts and exclusion of this exon in SMN2 transcripts are caused by the single nucleotide difference at +6 in SMN1 exon 7 (c.840C>T). Although the C to T change in SMN2 exon 7 does not change an amino acid, it does disrupt an exonic splicing enhancer (ESE) or creates an exon silencer element (ESS) that results in the majority of transcripts lacking exon 7.28,29 The ESEs and ESSs are cis-acting exonic sequences that influence the use of flanking splice sites. ESEs stimulate splicing and are often required for efficient intron removal, whereas ESSs inhibit splicing. Whether it is the loss of an ESE or creation of an ESS, the result is a reduction of full-length transcripts generated from SMN2. A single SMN2 gene produces less functional protein compared with a single SMN1 gene.9,10,20–30 Therefore, SMA arises because the SMN2 gene cannot fully compensate for the lack of functional SMN when SMN1 is mutated. However, small amounts of full-length transcripts generated by SMN2 are able to produce a milder type II or III phenotype when the copy number of the SMN2 gene is increased. SMA is, therefore, caused by low levels of SMN protein, rather than a complete absence of the protein. A recent report described three unrelated patients with SMA who possessed SMN2 copy numbers that did not correlate with the observed mild clinical phenotypes.31 A single base substitution in SMN2, c.859G>C, was identified in exon 7 in the patients DNA, and it was shown that the substitution created a new ESE element. The new ESE increased the amount of exon 7 inclusion and full-length transcripts generated from SMN2, thus resulting in the less severe phenotypes. Therefore, the SMA phenotype may not only be modified by the number of SMN2 genes but SMN2 sequence variations can also affect the disease severity. It should, therefore, not be assumed that all SMN2 genes are equivalent and sequence changes found within the SMN2 gene must be further investigated for potential positive or negative effects on SMN2 transcription when there is a lack of correlation between the genotype and phenotype.
Listing of mutations
Although the absence of both copies of the SMN1 gene is a very reliable and sensitive assay for the molecular diagnosis of SMA, approximately 5% of affected patients have other types of mutations in the SMN1 gene that will not be detected by homozygous deletion testing. Because of the high deletion frequency and according to the Hardy-Weinberg equilibrium, most of these patients will be compound heterozygotes; with one SMN1 allele being deleted and the other allele with a point mutation or other types of small mutations. If a patient with SMA possesses only a single copy of SMN1, it is likely that the remaining copy contains a more subtle mutation, including nonsense mutations, missense mutations, splice site mutation insertions, and small deletions. Many of the same intragenic mutations have now been reported in unrelated patients.19,32,33 The most frequently reported mutations are the p.Tyr272Cys (c.815A>G), c.399_402delAGAG, c.770-780dup11, and p.Thr274Ile (c.821C>T). The coding region for SMN can be found on ensemble (www.ensemble.org). Proper nomenclature must be used to report any verified sequence mutation (http://www.hgvs.org/mutnomen/).
Ethnic association of SMA
With an incidence of approximately 1 in 10,000 live births and a carrier frequency of 1/40–1/60, SMA is the leading inherited cause of infant mortality. A recent report provides carrier frequencies in several populations including white, Ashkenazi Jewish, African American, Asian, and Hispanic.34 The lower carrier frequencies found in African Americans and Hispanics would suggest a lower prevalence of SMA in these populations.
GUIDELINES
Definition of normal and mutation category
Alleles
Among normal alleles, a chromosome with one copy of SMN1 gene is designated as a “1-copy” or “1” allele. A chromosome bearing two copies of SMN1 gene is designated as a “2-copy” or “2” allele. The most common disease allele bears a deletion (or gene conversion of SMN1 to SMN2) resulting in a loss of exon 7 (Δ7SMN1) is referred to as the “0-copy” or “0” allele. Disease alleles with subtle intragenic point mutations on the SMN1 gene are referred to as 1d. The 0, 1, 2, and 1d alleles are also variably referred to as a, b, c, and d alleles, respectively, in literature.35 The resulting allele pairings that give rise to the diagnostic and carrier genotypes of SMA are defined later.
Diagnostic
The term “diagnostic” in the context of SMA is characterized by the presence of mutations classified into either of three categories (Table 1). Category 1, which accounts for approximately 95% of affected individuals, is characterized by a homozygous absence of SMN1 exon 7 (Δ7SMN1) due to deletions or gene conversions of SMN1 gene to SMN2 and is designated as the [0 + 0] genotype or a2.36 The resulting diagnostic finding is the absence of detectable SMN1 exon 7 and SMN1 exon 7 copy number of 0. Category 2, which accounts for approximately 5% of affected individuals, is characterized by compound heterozygosity for a rare intragenic point mutation within the SMN1 gene on one chromosome and a deletion/gene conversion of SMN1 exon 7 (Δ7SMN1) on the other chromosome and is designated as the [1d + 0] genotype or 2ad. The resulting diagnostic finding may be a detectable level of SMN1 exon 7 and an SMN1 exon 7 copy number of either 0 or 1 depending on the location of the rare intragenic point mutation within the SMN1 gene and its ability to interfere with SMN1 copy number analysis. Category 3, which is very rare and is most likely due to consanguinity, is characterized by subtle intragenic point mutations within the SMN1 gene on both chromosomes and is designated as the [1d + 1d] genotype or d2. The resulting diagnostic finding may be a detectable level of SMN1 exon 7 and an SMN1 exon 7 copy number of 0, 1, or 2 depending on the location of each rare intragenic point mutation within the SMN1 gene and its ability to interfere with SMN1 copy number analysis.
Carrier
The term “carrier” in the context of SMA is classified into either of three categories (Table 2). Category 1 is characterized by an SMN1 exon 7 copy number of 1 and presumes the presence of an SMN1 deletion/gene conversion on the other chromosome (heterozygous Δ7SMN1) and is designated as the [1 + 0] genotype or 2ab. Category 2 is characterized by a presence of two SMN1 genes in cis on a single chromosome along with a deletion/gene conversion of SMN1 exon 7 on the opposite chromosome resulting in an SMN1 exon 7 copy number of 2 and is designated as the [2 + 0] genotype or 2ac. Category 3 is characterized by a subtle intragenic point mutation on one chromosome resulting in an SMN1 exon 7 copy number of ≥2 and is designated as the [1 + 1d], and [2 + 1d] genotypes or 2bd and 2cd, respectively. It must be recognized that other rare carrier genotypes such as [3 + 0] or [3 + 1d] are likely and are thought to occur at a lower frequency relative to the most common carrier genotypes.36
Negative result
A negative test result is characterized by the presence of detectable amounts of SMN1 exon 7, with an SMN1 exon 7 copy number of >1, with the presence of subtle intragenic point mutations within the SMN1 gene having been ruled out. If the presence of subtle intragenic mutations has not been ruled out, a negative test result decreases the likelihood but does not exclude the diagnosis of SMA. Within the context of carrier testing, an SMN1 copy number of ≥2 is associated with a reduced risk to be a carrier.
TESTING CONSIDERATIONS
As with any genetic testing modality, the required intake information needed to facilitate an accurate result interpretation includes the reason for referral, i.e., diagnostic versus carrier testing, an accurate representation of family history, i.e., additional affected or carrier individuals identified within the family, clinical or diagnostic findings of relevance to SMA, and patient ethnicity. Patient ethnicity helps to provide appropriate risk assessment after a negative carrier testing result in an individual with no family history of SMA.
Diagnostic mutation analysis
Testing by SMN1 deletion or copy number analysis is indicated for individuals with a suspected diagnosis of SMA, presenting with symptoms of proximal muscle weakness, fasciculations, dysphagia, dysarthria, and absent deep tendon reflexes.
Sensitivity and specificity
The analytical sensitivity of SMN1 deletion/copy number analysis (proportion of homozygous Δ7SMN1 among all mutations in diagnostic category 1) is >99% (using the dosage assays described in the guidelines). The clinical sensitivity (proportion of homozygous Δ7SMN1 if 5q13-linked SMA is present) of the diagnostic test is approximately 95%. The remaining 5% of patients fall into either of diagnostic categories 2 or 3 (i.e., the [1d + 0] and the [1d + 1d] genotypes) and represent a source of false-negative diagnostic test results.3 These are patients with other types of small mutations within the SMN1 gene and will not be detected by the deletion testing.
Although the absence of both copies of the SMN1 gene is a very reliable and sensitive assay for the molecular diagnosis of SMA, approximately 5% of affected patients have other types of mutations in the SMN1 gene that will not be detected by homozygous deletion testing. Because of the high deletion frequency and according to the Hardy-Weinberg equilibrium, most of these patients will be compound heterozygotes; with one SMN1 allele being deleted and the other allele with a point mutation or other types of small mutations. If a patient with an SMA-like phenotype possesses only a single copy of SMN1, it is likely that the remaining copy contains a more subtle mutation, including nonsense mutations, missense mutations, splice site mutation insertions, and small deletions.
Both the analytical specificity (proportion of negative test results if homozygous Δ7SMN1 SMA genotype is not present) and the clinical specificity (proportion of negative test results if 5q13-linked SMA is not present) of SMN1 deletion/copy number analysis are >99%. Polymorphisms or point mutations under the primer and/or the probe binding regions may influence the analytical and clinical specificity by increasing the false-positive rate depending on the technology used.37 As a measure of additional quality assurance, follow-up sequencing underneath the primer and probe binding regions on all diagnostic (0 or 1 SMN1 copy number) results is expected to rule out a false-positive diagnostic finding attributable to this phenomenon and provide a better understanding of the underlying molecular mechanism of the mutation identified.
De novo deletion or gene conversions of paternal origin have been reported to occur at a frequency of 2% of patients with SMA.38 Carrier testing on both parents of patients with homozygous Δ7SMN1 SMA may provide additional information on the occurrence of de novo deletions. If one of the parents seems to be a noncarrier of SMA (i.e., ≥2 copies of SMN1), further carrier testing on both parents of the noncarrier parent (i.e., the grandparents) can be pursued to determine the phase of SMN1 genes in the noncarrier parent. This helps distinguish a [1 + 1] genotype leading to a de novo deletion in the index case, from a [2 + 0] obligate carrier genotype in the seemingly noncarrier parent. Furthermore, germline mosaicism for SMN1 deletion/gene conversions has been reported.6 Although the detection of a de novo mutation in an SMA family substantially lowers the recurrence risk, prenatal diagnosis in subsequent pregnancies should still be considered due to the rare possibility of a recurrent de novo mutation or germ-line mosaicism leading to an affected child.
Carrier testing
Carrier testing for SMA should be offered to asymptomatic individuals with a confirmed or suspected family history of SMA. Given the 1/40–1/60 carrier frequency of SMA, population carrier screening has recently been recommended by the ACMG39 but has not been supported by the American College of Obstetricians and Gynecologists' Committee on Genetics.40 Carrier frequencies determined from derived allele frequencies should consider all combinations of the most likely allele pairings expected to occur among carriers in the general population, namely [1 + 0], [1 + 1d], [2 + 0], and [2 + 1d]. Carrier frequencies derived solely from the observed 1-copy genotype frequencies tend to underestimate the prior risk estimates for carriers of SMA in the general population, thereby leading to inaccuracies in posterior risk estimates after carrier testing. The report by Ogino and Wilson35 provides Bayesian risk calculations for several situations.
The sensitivity (true positives/true positives + false negatives, or the detection rate) of the carrier test is defined as the proportion of carriers with the heterozygous Δ7SMN1, 1-copy genotype among all SMA carriers (including point mutation carriers and deletion carriers with two copies of the SMN1 gene) and can be expressed as the ratio [1 + 0]/[1 + 0] + [1 + 1d] + [2 + 0] + [2 + 1d]. Here a subset of the denominator, [1 + 1d] + [2 + 0] + [2 + 1d], represents a source of false-negative carrier test results. Detection rate varies by ethnicity ranging from 71% in African Americans to 95% in whites.34 The major contributor to this ethnicity-based variation in detection rate is the occurrence of two (or more) SMN1 genes in tandem on a single chromosome 5 (i.e., the [2 + 0] Category 2 carrier genotype). The estimated frequency of alleles with two or more copies of SMN1 is 3–8 times more prevalent in African Americans, when compared with other ethnic groups.34 This translates to a much higher frequency of individuals with the SMA carrier [2 + 0] genotype among African Americans compared with other races. It also has important implications in risk assessment and counseling after carrier screening in individuals of African American ancestry.
As with the diagnostic test, analytical specificity of the carrier test (proportion of negative test results if not a carrier) is >99% if the presence of polymorphisms underneath the primer and/or probe binding sites are ruled out. The negative predictive value, i.e., the proportion of negative tests that correctly identifies noncarriers (true negatives/true negatives + false negatives), is >99% regardless of ethnicity. The positive predictive value, i.e., the proportion of positive tests that correctly identify carriers of 5q13 linked SMA (true positive/true positive + false positives) is >99% if the presence of polymorphisms underneath the primer and/or probe binding sites are ruled out.
A negative test result within the context of carrier screening is defined by an SMN1 exon 7 copy number ≥2 with a reduced posterior risk for being a carrier. The posterior risk determination takes into consideration the conditional probability of being either a carrier or a noncarrier after an SMN1 copy number analysis result of ≥2.36,41 The posterior residual risk for an individual with no family history of SMA to be a carrier following the identification of two copies of SMN1 exon 7 varies by ethnicity ranging from 1:632 for an individual of white ancestry to 1:121 for an individual of African American ancestry.34 The residual risk following identification of three copies of SMN1 exon 7 is considerably lower and varies from 1:3500 for an individual of white ancestry to 1:3000 for an individual of African American ancestry.34 As with other carrier screening tests, patients need to understand that a negative carrier screen reduces but does not eliminate the risk to be a carrier of SMA. Risk assessment calculations using Bayesian analysis are essential for the proper genetic counseling of SMA families.35
The laboratory must establish validated, nonoverlapping cutoff values that can accurately and reliably distinguish SMN1 copy numbers of 0, 1, 2, and ≥3. The accuracy, precision, and confidence of SMN1 copy number measurements around these established cutoff values should be known to the laboratory. Copy number variations within the genomic internal standard and inefficiency of polymerase chain reaction (PCR) amplification of the internal standard reference gene relative to the SMN1 gene represent additional sources of false positive or incorrect copy number estimates. Therefore, the copy number of genomic internal standard reference gene should be constant at two copies within the genome, and the PCR amplification efficiency of the SMN1 gene relative to the chosen internal reference standard gene should be consistent between analyses. Performing replicate copy number measurements with two independent two copy number internal standard reference genes can help assure the accuracy of copy number analysis.
Prenatal testing
Indications for prenatal diagnosis of SMA include a 25% risk for the fetus to be affected (when both carrier parents are identified as a result of family history or following carrier identification by population screening) or the presence of abnormal findings such as decreased fetal movements and contractures in utero or increased NT on fetal ultrasound. Prenatal testing for SMA is performed by direct determination of the homozygous exon 7 deletion. A prerequisite is the previous identification of the homozygous deletion in the index case or positive carrier status in the parents. Testing both parents and the prenatal specimen by the same methodology, in addition to ruling out false-positive results attributed to sequence variants underneath the primer and probe binding sites, facilitates the most accurate interpretation of the prenatal test result. As maternal cell contamination of the fetal specimen can result in a false-negative test result, such contamination must be concurrently ruled out before reporting the prenatal test result. Although the presence of homozygous Δ7SMN1 is consistent with a diagnosis of SMA, the clinical severity, i.e., the type of SMA, cannot be predicted based on these molecular results.
Linkage
In families where a sample from a previous affected child is available, linkage analysis using microsatellite markers flanking the SMN1 gene may be considered when SMN1 deletion analysis is negative, and the presence of subtle intragenic mutations tracking within the family is suspected. Another indication for linkage analysis is in distinguishing a [1 + 1] genotype from a [2 + 0] genotype, when two copies of SMN1 are identified in the parent of an affected child with a homozygous Δ7SMN1. In this latter scenario, linkage analysis can also identify recombination events associated with de novo deletions occurring within the SMN1 gene.
SMN2 copy number
SMN2 copy number analysis is not routinely performed within the setting of diagnostic or carrier testing for SMA. SMN2 copy number analysis may be of value within the setting of clinical trials and newborn screening in stratifying patients who are more likely to respond to therapeutic strategies aimed at upregulating the levels of expression of full-length SMN protein from the SMN2 gene. Within clinical or prenatal diagnostic settings, however, results from SMN2 copy number analysis if available must be interpreted with caution. Although SMN2 copy number influences disease severity, SMN2 sequence variants and other genes have also been implicated in influencing SMA phenotype.42 Therefore, SMN2 copy number results may provide probabilistic information regarding clinical severity for an affected child or fetus but should not be viewed as definitive. Furthermore, an SMN2 copy number result indicates total SMN2 copy number for both alleles; therefore, it is not possible to determine SMN2 phase in unaffected individuals. For indications of carrier testing, SMN2 copy number determination does not provide information useful for counseling. For example, an SMN1 deletion carrier ([1 + 0] genotype) may carry three copies of SMN2, but it is not possible to determine how many of those SMN2 copies are in cis with the deletion and would, therefore, be passed to offspring. An ability to determine phase is not clinically available at this time but would be necessary to add value to determination of SMN2 copy number in carrier parents.
METHODOLOGICAL CONSIDERATIONS
Individual US laboratories offering molecular diagnostic and carrier testing for SMA should be in compliance with all federal and state regulations relevant to clinical laboratory operations. This includes meeting all CLIA/CAP quality control requirements. In addition, all laboratories should be active participants in annual CAP SMA proficiency testing challenges. All methodological applications should also be in compliance with the current Standards and Guideline for Clinical Genetics Laboratories developed by the Laboratory Practice Committee of the ACMG. Non-US laboratories should be similarly compliant with their individual countries statutory regulations governing oversight of clinical laboratories.
The absence of detectable SMN1 in patients with SMA is being used as a reliable and powerful diagnostic test for the majority of patients with SMA. The first diagnostic test for a patient suspected to have SMA should be the SMN1 gene deletion test. Both copies of the SMN1 exon 7 are absent in approximately 95% of affected patients, whereas small more subtle mutations have been identified in the remaining affected patients. The molecular diagnosis of the SMA consists of the detection of the absence of exons 7 of the SMN1 gene. Genetic testing is not only the most rapid and sensitive method to confirm the diagnosis but also the testing allows for further invasive investigations such as electromyography and muscle biopsy to be avoided. SMN1 dosage testing is used to determine the SMN1 copy number and detect SMA carriers: carriers will possess one SMN1 copy and noncarriers will have two SMN1 copies and occasionally have three SMN1 copies. There are a number of methods being used for the determination of SMN1 copy number, with multiplex ligation-dependent probe amplification (MLPA) and quantitative PCR (qPCR) being the most common. SMN1 sequencing is used for the identification of the compound heterozygote affected state. All general guidelines for PCR, restriction fragment length polymorphism (RFLP), MLPA, qPCR, and DNA sequencing in the ACMG Standards and Guidelines apply. These technologies have limitations and strengths when applied to diagnostic and/or carrier testing for SMA as detailed later.
RFLP test
The RFLP test is commonly used and allows for the detection of the homozygous deletion of SMN1 exon 7. It is currently being used as an assay for the diagnosis of SMA in both clinical and prenatal settings.43 The PCR primer sets first amplify both SMN1 and SMN2 exon 7. Although this is a highly repetitive region, the exon 7 base pair difference (840C>T) alters a DraI restriction enzyme site (due to a mismatched primer) and allows one to easily distinguish SMN1 from SMN2 on digestion of the PCR products. The absence of the undigested SMN1 exon 7 product is consistent with the SMA diagnosis. Presence of an undigested product band indicates one or more SMN1 copies. Another restriction enzyme that can be used in the RFLP assay is HinfI.19 In the HinfI assay, internal control restriction sites are introduced, which allows for the assessment of complete digestion. Regardless of the enzyme used, the appropriate positive and negative controls should always be included in every assay. Technical advantages of SMA RFLP include (1) reliability of the assay, (2) very robust and minimally sensitive to DNA quality or degradation, and (3) simplicity to set up and operate in a clinical diagnostic laboratory. Technical disadvantages of SMA RFLP include (1) the need to avoid partial digestion problems and (2) DNA sequence variants located under the SMN1 primer binding sites or enzymatic restriction site that may prevent the primers from annealing properly or proper digestion and might yield a false-positive diagnostic (zero copies) result. Clinical disadvantages of SMA RFLP include the inability to detect carrier status and determine SMN2 copy number.
Multiplex ligation-dependent probe amplification
MLPA is a PCR-based method of quantifying multiple genomic loci in a single reaction. It is based on the ligation of a set of two oligonucleotides probes that have annealed adjacently to a target sequence. Only ligated probes can serve as a template for a subsequent PCR. MLPA analysis of DNA from different individuals should reproducibly generate each of the expected peaks, and their sizes should correspond to those listed for each probe pair. MLPA technology is able to detect copy number of specific genomic loci and, therefore, can be used to test for SMA diagnostic and carrier status in clinical and prenatal settings.44 Probe sets specific to exon 7 of SMN1, SMN2 (MRC Holland SALSA MLPA KIT P021-A1 SMN Exon 7 probes 1260-L0966 and 1260-L0967), and 20 typically diploid loci located throughout the genome are used in SMA MLPA. After performing the enzymatic reactions, the PCR products are run through a capillary electrophoresis (CE) analyzer. The relative intensities and morphologies of the peaks should be consistent from one sample to the next. When the larger amplification products are weaker than the smaller fragments, this usually indicates that PCR amplification was not optimal or that the DNA sample analyzed might be degraded. Computer-aided scoring is a sensitive method to normalize the peak height or area for each PCR product compared with the 20 endogenous control loci. The median ratios across all the samples for each probe can be used as a reference value. Reference values for a copy number of 2 should approximately 1. A heterozygous deletion should give a ratio of approximately 0.5, whereas an elevated copy number should give a reference value >1.5. Reference values for a homozygous deletion (SMA positive) are close to 0. MLPA technology has several key advantages, including (1) allowance for simultaneous detection of SMN1 and SMN2 copy numbers, therefore, helping differentiate SMA type 1 from SMA types 2 and 3, (2) a high degree of precision for the quantitative detection of three or fewer SMN1 copies, (3) 20 independent control loci are able to be assayed in one reaction, (4) all reactions are performed in a single tube, and (5) probe sets are easy to obtain commercially, (6) a high degree of reproducibility and a large number of samples can be tested simultaneously, and (7) only 20 ng of genomic DNA is required, but 5–6-fold more DNA can be successfully technically validated. MLPA technology has several important limitations including (1) DNA sequence variants located under probe binding sites of SMN1 alleles may interfere with probe hybridization and might result in a false-positive carrier (one copy) or false-positive diagnostic (zero copies) result, (2) reactions are sensitive to contaminants but generate uninterpretable results, (3) MLPA cannot yet be used to investigate single cells, which is important for preimplantation genetic diagnosis testing, (4) MLPA is not a suitable method to detect unknown point mutations, (5) MLPA probes are sensitive to small deletions, insertions, and mismatches, and (6) MLPA requires a CE analyzer, which is a higher cost option compared with slab gel electrophoresis for RFLP.
Quantitative PCR
qPCR is a multiplex qPCR that coamplifies (in the same tube) multiple genomic loci to determine gene copy number.18 The multiplex can consist of competitive coamplification of SMN1, SMN2, the SMN internal standard, calibration factor, and the calibration factor internal standard. In the competitive PCR method, a known number of copies of a synthetic mutated internal standard are introduced with the patient sample into the PCR mixture. The internal standards are designed to be amplified with the same primer pairs for the SMN1 copy, with efficiencies similar to those of the genomic DNA counterparts and yield PCR products slightly smaller than the SMN PCR product. The copy number of SMN1 is determined by coamplification of SMN1, SMN2, and the internal standards, and the ratios are quantitated. End point detection of amplification of fluorescently tagged PCR products is done by running the samples through a CE analyzer. The major advantage of this technique is that the internal standard is amplified with the same primers that amplify the target sequence. Thus, the efficiency of the amplification of the patient DNA and the internal standard DNA should be very similar and allow one to accurately determine the gene copy number. Alternatively, real-time detection45 (as opposed to end point detection) of multiplex PCR reactions using hydrolysis or hybridization fluorescent probes uses other strategies for normalization including (1) standard curve method or (2) comparative threshold method. Using the standard curve method, a standard curve is first constructed from a DNA of known copy number. This curve is then used as a reference standard for extrapolating quantitative information for the SMN1 copy number. The cycle at which the curve crosses a specified threshold is called the cycle threshold (Ct). Variation introduced due to variable DNA inputs can be corrected by normalizing to the calibration factor. Using the comparative threshold method, the Ct values of the sample of interest are compared with the Ct values of the calibration factor. Ct values of both the calibrator and the sample of interest are normalized to an appropriate endogenous internal control gene. qPCR is able to detect copy number of specific genomic loci and, therefore, can be used to test for SMA diagnostic and carrier status. Key advantages of qPCR include (1) extreme sensitivity, allowing the detection of less than five copies (perhaps only one copy in some cases) of a target sequence, making it possible to analyze small samples such as single cell analysis for the purpose of preimplantation genetic diagnosis, (2) with appropriate internal standards and calculations, mean variation coefficients are 5–10%, allowing reproducible analysis of the gene copy number, (3) all real-time platforms are relatively quick, with some affording high-throughput automation, and (4) real-time platforms are performed in a closed reaction vessel that requires no post-PCR manipulations, thereby minimizing the chances for cross contamination in the laboratory. Important limitations for qPCR techniques include (1) compounds present in certain biological samples or sample collection compounds (i.e., heparin) may inhibit PCR, (2) DNA sequence variants located under the primer binding sites of primers may reduce proper annealing and might result in a false-positive carrier (one copy) or diagnostic (zero copies) result, and (3) improper assay development and incorrect data analysis. Unwarranted conclusions may present the largest limitation of qPCR, and therefore, a robust and extensive validation is warranted to ensure specificity and accuracy of the results. Amplification and melting curves must be visually inspected, whereas independent calculations based on these curves should be double checked for accuracy. Finally, neither the MLPA nor qPCR can determine whether two SMN1 genes are in cis on a single chromosome.
DNA sequencing
Although the absence of both copies of the SMN1 gene is a very reliable and sensitive assay for the molecular diagnosis of SMA, approximately 5% of affected patients have other types of mutations in the SMN1 gene that will not be detected by homozygous deletion testing. Because of the high deletion frequency and according to the Hardy-Weinberg equilibrium, most of these patients will be compound heterozygotes; with one SMN1 allele being deleted and the other allele with a point mutation or other types of small mutations. If a patient with a SMA-like phenotype possesses only a single copy of SMN1, it is likely that the remaining copy contains a more subtle mutation, including nonsense mutations, missense mutations, splice site mutation insertions, and small deletions. The development of high-throughput DNA sequencing techniques has made direct DNA sequencing of PCR-amplified genomic DNA a rapid and economical approach to the identification of sequence mutations. As a consequence of the SMN1 gene being relatively small and given the uniform spectrum of mutations, it is a relatively straightforward procedure to sequence the gene and identify mutations in patients who are negative for the diagnostic deletion test. However, it is necessary to verify that the intragenic mutation has occurred in the SMN1 gene and not the SMN2 gene. As an initial screen, primers that do not distinguish between SMN1 and SMN2 may be used to amplify each exon for direct DNA sequencing. If variants or mutations are identified, SMN1-specific long-range PCR amplification is followed by either direct DNA sequencing of that long-range product or nested PCR sequencing.46 Important limitations for SMN direct gene sequencing include (1) the requirement of allele-specific sequencing of all variants identified, (2) DNA sequencing does not detect large deletions or insertions, (3) mutations in patients exhibiting mosaicism or chromosomal rearrangements may not be detectable using sequencing technology, and (4) variants of unknown significance.
INTERPRETATIONS
Elements considered essential to the reporting of clinical test results are described in detail in the current ACMG Standards and Guidelines for Clinical Genetics Laboratories. Examples of model laboratory reports are included in the appendix. The following elements must also be included in the reporting of SMA results.
The methodology used to assign the SMA genotype should be clearly stated. All positive results in clinically diagnosed/suspected individuals should state that genetic counseling is indicated, and carrier testing is available for other at-risk family members.
Comments on phenotype, if included, should be abstract rather than case specific. Although the inverse relationship between the SMN2 copy number and disease severity has been well established, the report should clearly state that the relationship is not absolute if reporting SMN2 levels. It is important that couples undergoing carrier screening recognize that the carrier test does not provide genotype/phenotype information. Type I SMA occurs in approximately 60% of the cases, whereas the milder types II and III account for the remaining 40% of the cases.
Alternative diagnosis may be included when two normal copies of the SMN1 gene have been detected. Other motor neuron disorders should be considered such as SMA with respiratory distress, X-linked SMA, distal muscular atrophy, and juvenile amyotrophic lateral sclerosis.
It is imperative that individuals understand the limitations of the carrier test: two SMN1 genes in cis on the one chromosome 5, presence of rare de novo mutations, and the nondeletion mutations. The issue of these false-negative results must be included on all negative carrier reports. As is true for carrier screening programs, the testing must be voluntary, and assurance of confidentiality is absolutely necessary.
Informed consent and the usual caveats should be addressed including paternity issues, possible diagnostic errors due to sample mix-ups, and genotype errors due the presence of rare polymorphisms.
The following statement must be included on the report: “This test was developed and its performance characteristics determined by this laboratory. It has not been cleared or approved by the US Food and Drug Administration (FDA). The FDA has determined that such clearance or approval is not necessary. This analysis is used for clinical purposes. It should not be regarded as investigational or for research.”
References
Pearn J . Incidence, prevalence, and gene frequency studies of chronic childhood spinal muscular atrophy. J Med Genet 1978; 15: 409–413.
Munstat TL, Davies KE . International SMA consortium meeting. Neuromuscular Disord 1992; 2: 423–428.
Zerres K, Rudnik-Schoneborn S . Natural history in proximal spinal muscular atrophy: clinical analysis of 445 patients and suggestions for a modification of existing classifications. Arch Neurol 1995; 52: 518–523.
Meldrum C, Scott C, Swoboda KJ . Spinal muscular atrophy genetic counseling access and genetic knowledge: parents perspective. J Child Neurol 2007; 22: 1019–1026.
Lefebvre S, Burglen L, Reboullet S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995; 80: 155–165.
Burglen L, Lefebvre S, Clermont O, et al. Structure and organization of the human survival motor neuron (SMN) gene. Genomics 1996; 32: 479–482.
Monani UR, McPherson JD, Burghes AH . Promoter analysis of the human centromeric and telomeric survival motor neuron genes (SMNC and SMNT). Biochim Biophys Acta 2007; 1445: 330–336.
Echaniz-Laguna A, Miniou P, Bartholdi D, Melki J . The promoters of the survival motor neuron gene (SMN) and its copy (SMNc) share common regulatory elements. Am J Hum Genet 1999; 64: 1354–1370.
Lefebvre S, Burlet P, Liu Q, et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet 1997; 16: 265–269.
Coovert DD, Le TT, McAndrew PE, et al. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet 1997; 6: 1205–1214.
Paushkin S, Gubitz AK, Massenet S, Dreyfuss G . The SMN complex, an assemblyosome of ribonucleoproteins. Curr Opin Cell Biol 2002; 14: 305–312.
Yong J, Wan L, Dreyfuss G . Why do cells need an assembly machine for RNA-protein complexes?. Trends Cell Biol 2004; 14: 226–232.
Monani UR . Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron 2005; 48: 885–896.
Zhang HL, Pan F, Hong D, Shenoy SM, Singer RH, Bassell GJ . Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J Neurosci 2003; 23: 6627–6637.
Rossoll W, Jablonka S, Andreassi C, et al. SMN, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J Cell Biol 2003; 163: 801–812.
Wirth B . An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy. Hum Mutat 2000; 15: 228–237.
Campbell L, Potter A, Ignatius J, Dubowitz V, Davies K . Genomic variation and gene conversion in spinal muscular atrophy: implications for disease process and clinical phenotype. Am J Hum Genet 1997; 61: 40–50.
McAndrew PE, Parsons DW, Simard LR, et al. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet 1997; 60: 1411–1422.
Wirth B, Herz M, Wetter A, et al. Quantitative analysis of survival motor neuron copies: identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype-phenotype correlation, and implications for genetic counseling. Am J Hum Genet 1999; 64: 1340–1356.
Mailman MD, Heinz JW, Papp AC, et al. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet Med 2002; 4: 20–26.
Prior TW, Swoboda KJ, Scott HD, Hejmanowski AQ . Homozygous SMN1 deletions in unaffected family members and modification of the phenotype by SMN2. Am J Med Genet A 2004; 130: 207–310.
Hsieh-Li HM, Chang JG, Jong YJ, et al. A mouse model for spinal muscular atrophy. Nat Genet 2000; 26: 66–70.
Monani UR, Coovert DD, Burghes AH . Animal models of spinal muscular atrophy. Hum Mol Genet 2000; 9: 2451–2457.
Hoffman Y, Lorson CL, Stamm S, Androphy EJ, Wirth B . Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2). Proc Natl Acad Sci USA 2000; 97: 9618–9623.
Oprea GE, Krober S, McWhorter ML, et al. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science 2008; 320: 524–527.
Lorson CL, Androphy EJ . An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum Mol Genet 2000; 9: 259–265.
Lorson CL, Hahnen E, Androphy EJ, Wirth B . A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA 1999; 96: 6307–6311.
Cartegni L, Kraniner AR . Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet 2002; 30: 377–384.
Kashima T, Manley JL . A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet 2003; 34: 460–463.
Gavrilov DK, Shi X, Das K, Gilliam TC, Wang CH . Differential SMN2 expression associated with SMA severity. Nat Genet 1998; 20: 230–231.
Prior TW, Krainer AR, Hua Y, et al. A positive modifier of spinal muscular atrophy in the SMN2 gene. Am J Hum Genet 2009; 85: 408–413.
Alias L, Bernal S, Fuentes-Prior P, et al. Mutation update of spinal muscular atrophy in Spain: molecular characterization of 745 unrelated patients and identification of four novel mutations in the SMN1 gene. Hum Genet 2009; 125: 29–39.
Parsons DW, McAndrew PE, Iannaccone ST, et al. Intragenic telSMN mutations: frequency, distribution, evidence of a founder effect and modification of spinal muscular atrophy phenotype by cenSMN copy number. Am J Hum Genet 1998; 63: 1712–1723.
Hendrickson BC, Donohoe C, Akmaev VR, et al. Differences in SMN1 allele frequencies among ethnic groups within North America. J Med Genet 2009; 46: 641–644.
Ogino S, Wilson RB . Genetic testing and risk assessment for spinal muscular atrophy (SMA). Hum Genet 2002; 111: 477–500.
Ogino S, Wilson RB, Gold B . New insights on the evolution of the SMN1 and SMN2 region: simulation and meta-analysis for allele and haplotype frequency calculations. Eur J Hum Genet 2004; 12: 1015–1023.
Eggermann T . A new splice site mutation in the SMN1 gene causes discrepant results in SMN1 deletion screening approaches. Neuromusc dis 2008; 18: 146–149.
Wirth B, Schmidt T, Hahnen E, et al. De novo rearrangements found in 2% of index patients with spinal muscular atrophy: mutational mechanisms, parental origin, mutation rate, and implications for genetic counseling. Am J Hum Genet 1997; 61: 1102–1111.
Prior TW . Carrier screening for spinal muscular atrophy. Genet Med 2008; 10: 840–842.
ACOG Committee Opinion Spinal muscular atrophy. Obstet Gynecol 2009; 113: 1194–1196.
Smith M, Calabro V, Chong B, et al. Population screening and cascade testing for carriers of SMA. Eur J Hum Genet 2007; 15: 759–766.
Prior TW . Perspectives and diagnostic considerations in spinal muscular atrophy. Genet Med 2010; 12: 145–152.
van der Steege G, Grootscholten P, van der Vlies, et al. PCR-based DNA test to confirm clinical diagnosis of autosomal recessive spinal muscular atrophy. Lancet 1995; 345: 985–986.
Huang C-H, Chang YY, Chen CH . Copy number analysis of survival motor neuron genes by multiplex ligation-dependent probe amplification. Genet Med 2007; 9: 241–248.
Anhuf D, Eggermann T, Rudnik-Schoneborn S, Zerres K . Determination of SMN1 and SMN2 copy number using Taqman technology. Hum Mutat 2003; 22: 74–78.
Cheng S, Fockler C, Barnes WM, Higuchi R, Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci USA 1994 91: 5695–5699.
Acknowledgements
The development of this laboratory practice guideline was supported by an educational Grant from the Claire Altman Heine Foundation, Inc., to the American College of Medical Genetics Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure: N.N. and E.A.S. are employees of Genzyme Genetics. S.D.B. and C.B. are employees of Athena Diagnostics. Both of these companies perform laboratory testing for SMA. T.W.P. does not have a commercial interest in SMA testing but does perform the testing in his laboratory. In addition, he has received generous research funding from the Claire Altman Heine Foundation, Inc. Genzyme Genetics and its logo are trademarks of Genzyme Corporation and used by Esoterix Genetic Laboratories, LLC, a wholly owned subsidiary of LabCorp, under license. Esoterix Genetic Laboratories and LabCorp are operated independently from Genzyme Corporation.
Disclaimer: These ACMG Standards and Guidelines are developed primarily as an educational resource for clinical laboratory geneticists to help them provide quality clinical laboratory genetic services. Adherence to these standards and guidelines is voluntary and does not necessarily assure a successful medical outcome. These Standards and Guidelines should not be considered inclusive of all proper procedures and tests or exclusive of other procedures and tests that are reasonably directed to obtaining the same results. In determining the propriety of any specific procedure or test, the clinical laboratory geneticist should apply his or her own professional judgment to the specific circumstances presented by the individual patient or specimen.Clinical laboratory geneticists are encouraged to document in the patient's record the rationale for the use of a particular procedure or test, whether or not it is in conformance with these Standards and Guidelines. They also are advised to take notice of the date any particular standard or guidelines was adopted, and to consider other relevant medical and scientific information that becomes available after that date. It also would be prudent to consider whether intellectual property interests may restrict the performance of certain tests and other procedures.
APPENDIX: MODEL LABORATORY REPORTS
APPENDIX: MODEL LABORATORY REPORTS
Indication
Screening/carrier test/negative family history.
Reported ethnicity
White.
Comment
SMA is an autosomal recessive disease of variable age of onset and severity caused by mutations (most often deletions or gene conversions) in the SMN1 gene. Molecular testing assesses the number of copies of the SMN1 gene. Affected individuals have 0 copies of the SMN1 gene. Individuals with one copy of the SMN1 gene are predicted to be carriers of SMA. Individuals with two or more copies have a reduced risk to be carriers. This copy number analysis cannot detect individuals who are carriers of SMA as a result of either two (or very rarely three) copies of the SMN1 gene on one chromosome and the absence of the SMN1 gene on the other chromosome or small intragenic mutations within the SMN1 gene. This analysis also will not detect germline mosaicism or mutations in genes other than SMN1. Additionally, de novo mutations have been reported in approximately 2% of patients with SMA.
Methods
Specimen DNA is isolated and amplified by real-time PCR for exon 7 of the SMN1 gene and two reference genes. A mathematical algorithm is used to calculate the number of copies of SMN1. Sequencing of the primer and probe binding sites for the SMN1 real-time PCR reaction is performed on all fetal samples and on samples from individuals with one copy of SMN1 on carrier testing, to rule out the presence of sequence variants, which could interfere with analysis and interpretation. This test was developed and its performance characteristics determined by this laboratory. It has not been cleared or approved by the FDA. The FDA has determined that such clearance or approval is not necessary. This analysis is used for clinical purposes. It should not be regarded as investigational or for research.
General disclaimer
False-positive or -negative results may occur for reasons that include genetic variants, blood transfusions, bone marrow transplantation, erroneous representation of family relationships, or contamination of a fetal sample with maternal cells.
Example of a negative SMA carrier report
Result
SMN1 copy number: 2 (reduced carrier risk).
Interpretation
This individual has an SMN1 copy number of 2. This result reduces but does not eliminate the risk to be a carrier of SMA. Ethnic-specific risk reductions based on negative family history and an SMN1 copy number of 2 are provided in the comment section. Genetic counseling is recommended.
Example of a positive SMA carrier report
Result
SMN copy number: 1 (carrier).
Interpretation
This individual has one copy of SMN1 and is, therefore, predicted to be a carrier of SMA, a disease of variable age of onset and severity. Genetic counseling is recommended.
Rights and permissions
About this article
Cite this article
Prior, T., Nagan, N., Sugarman, E. et al. Technical standards and guidelines for spinal muscular atrophy testing. Genet Med 13, 686–694 (2011). https://doi.org/10.1097/GIM.0b013e318220d523
Published:
Issue Date:
DOI: https://doi.org/10.1097/GIM.0b013e318220d523
Keywords
This article is cited by
-
Next generation sequencing is a highly reliable method to analyze exon 7 deletion of survival motor neuron 1 (SMN1) gene
Scientific Reports (2022)
-
Revealing diverse alternative splicing variants of the highly homologous SMN1 and SMN2 genes by targeted long-read sequencing
Molecular Genetics and Genomics (2022)
-
Gene selection for the Australian Reproductive Genetic Carrier Screening Project (“Mackenzie’s Mission”)
European Journal of Human Genetics (2021)
-
Development and validation of a 4-color multiplexing spinal muscular atrophy (SMA) genotyping assay on a novel integrated digital PCR instrument
Scientific Reports (2020)
-
SMN1 gene copy number analysis for spinal muscular atrophy (SMA) in a Turkish cohort by CODE-SEQ technology, an integrated solution for detection of SMN1 and SMN2 copy numbers and the “2+0” genotype
Neurological Sciences (2020)