Abstract
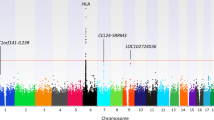
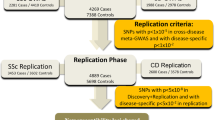
Sarcoidosis is a complex disease of unknown etiology characterized by the presence of granulomatous inflammation. Though various immune system pathways have been implicated in disease, the relationship between the genetic determinants of sarcoidosis and other inflammatory disorders has not been characterized. Herein, we examined the degree of genetic pleiotropy common to sarcoidosis and other inflammatory disorders to identify shared pathways and disease systems pertinent to sarcoidosis onset. To achieve this, we quantify the association of common variant polygenic risk scores from nine complex inflammatory disorders with sarcoidosis risk. Enrichment analyses of genes implicated in pleiotropic associations were further used to elucidate candidate pathways. In European-Americans, we identify significant pleiotropy between risk of sarcoidosis and risk of asthma (R2=2.03%; P=8.89 × 10−9), celiac disease (R2=2.03%; P=8.21 × 10−9), primary biliary cirrhosis (R2=2.43%; P=2.01 × 10−10) and rheumatoid arthritis (R2=4.32%; P=2.50 × 10−17). These associations validate in African Americans only after accounting for the proportion of genome-wide European ancestry, where we demonstrate similar effects of polygenic risk for African-Americans with the highest levels of European ancestry. Variants and genes implicated in European-American pleiotropic associations were enriched for pathways involving interleukin-12, interleukin-27 and cell adhesion molecules, corroborating the hypothesized immunopathogenesis of disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA 2009; 106: 9362–9367.
Sivakumaran S, Agakov F, Theodoratou E, Prendergast JG, Zgaga L, Manolio T et al. Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet 2011; 89: 607–618.
Ellinghaus D, Jostins L, Spain SL, Cortes A, Bethune J, Han B et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet 2016; 48: 510–518.
International Schizophrenia C, Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–752.
Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet 1998; 63: 1839–1851.
Iannuzzi MC, Rybicki BA, Teirstein AS . Sarcoidosis. N Engl J Med 2007; 357: 2153–2165.
Sverrild A, Backer V, Kyvik KO, Kaprio J, Milman N, Svendsen CB et al. Heredity in sarcoidosis: a registry-based twin study. Thorax 2008; 63: 894–896.
James DG . Descriptive definition and historic aspects of sarcoidosis. Clin Chest Med 1997; 18: 663–679.
Rybicki BA, Major M, Popovich J Jr, Maliarik MJ, Iannuzzi MC . Racial differences in sarcoidosis incidence: a 5-year study in a health maintenance organization. Am J Epidemiol 1997; 145: 234–241.
Edmondstone WM, Wilson AG . Sarcoidosis in Caucasians, Blacks and Asians in London. Br J Dis Chest 1985; 79: 27–36.
Rybicki BA, Hirst K, Iyengar SK, Barnard JG, Judson MA, Rose CS et al. A sarcoidosis genetic linkage consortium: the sarcoidosis genetic analysis (SAGA) study. Sarcoidosis Vasc Diffuse Lung Dis 2005; 22: 115–122.
Adrianto I, Lin CP, Hale JJ, Levin AM, Datta I, Parker R et al. Genome-wide association study of African and European Americans implicates multiple shared and ethnic specific loci in sarcoidosis susceptibility. PLoS ONE 2012; 7: e43907.
Fischer A, Ellinghaus D, Nutsua M, Hofmann S, Montgomery CG, Iannuzzi MC et al. Identification of immune-relevant factors conferring sarcoidosis genetic risk. Am J Respir Crit Care Med 2015; 192: 727–736.
Levin AM, Iannuzzi MC, Montgomery CG, Trudeau S, Datta I, Adrianto I et al. Admixture fine-mapping in African Americans implicates XAF1 as a possible sarcoidosis risk gene. PloS ONE 2014; 9: e92646.
Rivera NV, Ronninger M, Shchetynsky K, Franke A, Nothen MM, Muller-Quernheim J et al. High-density genetic mapping identifies new susceptibility variants in sarcoidosis phenotypes and shows genomic-driven phenotypic differences. Am J Respir Crit Care Med 2016; 193: 1008–1022.
Lareau CA, Adrianto I, Levin AM, Iannuzzi MC, Rybicki BA, Montgomery CG . Fine mapping of chromosome 15q25 implicates ZNF592 in neurosarcoidosis patients. Ann Clin Transl Neurol 2015; 2: 972–977.
Darlington P, Tallstedt L, Padyukov L, Kockum I, Cederlund K, Eklund A et al. HLA-DRB1* alleles and symptoms associated with Heerfordt's syndrome in sarcoidosis. Eur Respir J 2011; 38: 1151–1157.
Bello GA, Adrianto I, Dumancas GG, Levin AM, Iannuzzi MC, Rybicki BA et al. Role of NOD2 pathway genes in sarcoidosis cases with clinical characteristics of Blau syndrome. Am J Respir Crit Care Med 2015; 192: 1133–1135.
Thompson IA, Liu B, Sen HN, Jiao X, Katamay R, Li Z et al. Association of complement factor H tyrosine 402 histidine genotype with posterior involvement in sarcoid-related uveitis. Am J Ophthalmol 2013; 155: 1068–1074 e1.
Gialafos E, Triposkiadis F, Kouranos V, Rapti A, Kosmas I, Manali E et al. Relationship between tumor necrosis factor-alpha (TNFA) gene polymorphisms and cardiac sarcoidosis. In Vivo 2014; 28: 1125–1129.
Euesden J, Lewis CM, O'Reilly PF . PRSice: Polygenic Risk Score software. Bioinformatics 2015; 31: 1466–1468.
Power RA, Steinberg S, Bjornsdottir G, Rietveld CA, Abdellaoui A, Nivard MM et al. Polygenic risk scores for schizophrenia and bipolar disorder predict creativity. Nat Neurosci 2015; 18: 953–955.
Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013; 381: 1371–1379.
Rajoriya N, Wotton CJ, Yeates DG, Travis SP, Goldacre MJ . Immune-mediated and chronic inflammatory disease in people with sarcoidosis: disease associations in a large UK database. Postgrad Med J 2009; 85: 233–237.
Chung D, Yang C, Li C, Gelernter J, Zhao H . GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet 2014; 10: e1004787.
Zhang B, Kirov S, Snoddy J . WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 2005; 33 (Web Server issue): W741–W748.
Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R . ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res 2011; 39 (Database issue): D712–D717.
Moller DR, Forman JD, Liu MC, Noble PW, Greenlee BM, Vyas P et al. Enhanced expression of IL-12 associated with Th1 cytokine profiles in active pulmonary sarcoidosis. J Immunol 1996; 156: 4952–4960.
Larousserie F, Pflanz S, Coulomb-L'Hermine A, Brousse N, Kastelein R, Devergne O . Expression of IL-27 in human Th1-associated granulomatous diseases. J Pathol 2004; 202: 164–171.
Katchar K, Soderstrom K, Wahlstrom J, Eklund A, Grunewald J . Characterisation of natural killer cells and CD56+ T-cells in sarcoidosis patients. Eur Respir J 2005; 26: 77–85.
Rossman MD, Thompson B, Frederick M, Maliarik M, Iannuzzi MC, Rybicki BA et al. HLA-DRB1*1101: a significant risk factor for sarcoidosis in blacks and whites. Am J Hum Genet 2003; 73: 720–735.
Hofmann S, Franke A, Fischer A, Jacobs G, Nothnagel M, Gaede KI et al. Genome-wide association study identifies ANXA11 as a new susceptibility locus for sarcoidosis. Nat Genet 2008; 40: 1103–1106.
Oh SS, Galanter J, Thakur N, Pino-Yanes M, Barcelo NE, White MJ et al. Diversity in clinical and biomedical research: a promise yet to be fulfilled. PLoS Med 2015; 12: e1001918.
Hoffmann TJ, Kvale MN, Hesselson SE, Zhan Y, Aquino C, Cao Y et al. Next generation genome-wide association tool: design and coverage of a high-throughput European-optimized SNP array. Genomics 2011; 98: 79–89.
Vilhjalmsson BJ, Yang J, Finucane HK, Gusev A, Lindstrom S, Ripke S et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet 2015; 97: 576–592.
Dudbridge F . Power and predictive accuracy of polygenic risk scores. PLoS Genet 2013; 9: e1003348.
James DG, Sharma OP . Overlap syndromes with sarcoidosis. Postgrad Med J 1985; 61: 769–771.
Lowe G, Johnston RN . Sarcoidosis and coeliac disease. Lancet 1984; 2: 637.
Naseer T, Minshall EM, Leung DY, Laberge S, Ernst P, Martin RJ et al. Expression of IL-12 and IL-13 mRNA in asthma and their modulation in response to steroid therapy. Am J Respir Crit Care Med 1997; 155: 845–851.
Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet 2011; 43: 887–892.
Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM . Design and analysis of admixture mapping studies. Am J Hum Genet 2004; 74: 965–978.
Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet 2010; 42: 295–302.
Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015; 47: 979–986.
International Multiple Sclerosis Genetics C, Bush WS, Sawcer SJ, de Jager PL, Oksenberg JR, McCauley JL et al. Evidence for polygenic susceptibility to multiple sclerosis—the shape of things to come. Am J Hum Genet 2010; 86: 621–625.
Cordell HJ, Han Y, Mells GF, Li Y, Hirschfield GM, Greene CS et al. International genome-wide meta-analysis identifies new primary biliary cirrhosis risk loci and targetable pathogenic pathways. Nat Commun 2015; 6: 8019.
Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet 2010; 42: 508–514.
Bentham J, Morris DL, Cunninghame Graham DS, Pinder CL, Tombleson P, Behrens TW et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet 2015; 47: 1457–1464.
Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009; 41: 703–707.
Acknowledgements
We thank the OMRF Genomics core for performing the genotyping experiments and the useful discussion. We are also grateful to the EVE asthma analysis consortium (RC2 HL101651) and particularly the efforts of Drs Carole Ober, Dan Nicolae and Dara Torgerson to make the asthma summary statistics available to the broader scientific community. Research reported in this publication was supported by the National Institute of General Medical Sciences (NIGMS), Heart, Lung and Blood Institute (NHLBI), and National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health under award numbers: P20GM103456 and P30GM110766 to IA, R01-HL54306 and U01-HL060263 to MCI, R56-AI072727 and R01-HL092576 to BAR, and P30 GM110766-01, RC2HL101499 and R01HL113326 to CGM. CAL is supported by an NSF Graduate Research Fellowship # DGE1144152. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
Conceived and designed the study: CAL, CFD, MCI, BAR, AML and CGM. Analyzed data: CAL, CFD, IA and AML. Interpreted the data, drafted/revised the manuscript: CAL, CFD, IA, CJL, PMG, MCI, BAR, AML and CGM.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Genes and Immunity website
Supplementary information
Rights and permissions
About this article
Cite this article
Lareau, C., DeWeese, C., Adrianto, I. et al. Polygenic risk assessment reveals pleiotropy between sarcoidosis and inflammatory disorders in the context of genetic ancestry. Genes Immun 18, 88–94 (2017). https://doi.org/10.1038/gene.2017.3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2017.3