Abstract
Purpose
To investigate the possibility of collagen type VIII α2 (COL8A2) as a potential susceptibility gene for Korean patients with Fuchs' corneal dystrophy (FECD), we performed mutation screening of the COL8A2 gene.
Methods
A total of 25 FECD patients were screened, including 15 patients from six pedigrees with early onset FECD and an additional 10 unrelated patients, all of Korean ancestry. Seventy-three control individuals without corneal disease were selected from the general population. PCR—SSCP and direct sequencing were used to screen genetic variations in COL8A2. The pathogenic impact of these sequence variants was evaluated through the SIFT and PolyPhen algorithms.
Results
We have identified a novel heterozygous mutation, Q455V, in exon 2 of COL8A2. All patients of Korean pedigrees with FECD had the Q455V mutation, and two out of nine unrelated cases also had this mutation. But it was not present in unaffected individuals from these pedigrees or from control groups. Two heterozygous missense mutations, R155Q and T502M, were also observed, but, they showed no significant difference between FECD patients and controls. The allele frequencies of A35A and G495G, which were synonymous substitutions, were significantly associated with FECD. Both Q455V and T502M were predicted as deleterious mutations by computational methods using PolyPhen and SIFT.
Conclusions
Our data constitute the first report of a heterozygous Q455V mutation of the COL8A2 gene in Korean patients with FECD. Q455V may be the causative defect in the development and progression of Korean FECD patients.
Similar content being viewed by others
Introduction
Fuchs' corneal dystrophy (FECD) is a bilateral, progressive degenerative disease of corneal endothelial cells, which leads to progressive edema of the corneal stroma.1, 2 FECD can be characterized by the formation of guttae, microscopic refractile excrescences in the collagen-rich basal lamina secreted by the corneal endothelium. These structures block ion transport and solute barrier functions in the endothelial layer,3, 4 which leads to the progressive dysfunction of the corneal endothelium. People with this disease are currently the most frequent recipients of corneal transplants.5 FECD generally begins in the fifth decade of life and can progress slowly over the next two to three decades.6, 7 Although rare, there is also an early onset form of this disease.2, 7, 8
Although its pathogenesis has not been clearly elucidated, FECD is known to be an autosomal dominant disease with high penetrance that develops independently from systemic or environmental factors.7, 8, 9 Therefore, understanding the genetic component of this disease is critical for the development of future therapies and preventative care. Three FECD susceptibility genomic loci 1p34.3-32, 13q12.11-q12.13, and 18q21.2-q21.32 have been previously identified through linkage studies of multigenerational families with autosomal-dominant FECD.8, 10, 11 On the basis of studies reporting significant linkage of FECD, several pathogenic mutations in the collagen type VIII α2 gene (COL8A2), located on 1p34.3-p32, have been detected in affected individuals with FECD, and a strong association between FECD patients and genetic variants of COL8A2 has been validated by multiple studies.6, 7, 8, 12 In particular, two missense mutations, L450W and Q455K, showed perfect concordance in an FECD family having early onset and are positioned within the triple-helical domain of α2 collagen type VIII, which leads to the structural alteration of Descemet's membrane.6, 8, 13, 14 Also, in English FECD patients, three missense mutations of the COL8A2 gene, R155Q, R304Q, and R434H, were identified in familial and unrelated forms of common FECD,8 whereas Aldave et al (2006) and Kobayashi et al (2004) reported that no COL8A2 variants associated with the common FECD subtype, late-onset FECD have been identified.15 Furthermore, an observed association between the COL8A2 gene variants and FECD, as well as other corneal diseases, including keratoconus and posterior polymorphous corneal dystrophy (PPCD), was reported by others.6, 8, 12, 15 Therefore, it is important to identify the genetic factors that determine susceptibility to FECD to gain an insight into the pathogenesis of FECD in Korean patients, because the susceptibility of mutations in COL8A2 can vary in different ethnicities and in the age of onset.8, 12, 15, 16, 17 The association between variations in the COL8A2 gene and Korean FECD patients is yet to be investigated. In this study, we screened the COL8A2 gene for both familial and unrelated FECD patients to determine whether mutations in COL8A2 are responsible for causing FECD in a Korean population. In addition, we used two sequences homology-based software tools to predict the likely impact of the observed nonsynonymous mutations on the protein function.
Materials and methods
Subjects
A total of 25 FECD patients were screened, including 15 patients from six pedigrees with early onset FECD and an additional 10 unrelated patients, all of Korean ancestry. Each individual was examined and photographed using slit lamp biomicroscopy to document the presence of guttae. Seventy-three control individuals without corneal disease were selected from the general population. All FECD patients and controls were identified from the Korea Eye Tissue and Gene Bank related to Blindness, Department of Ophthalmology, the Catholic University of Korea. Appropriate informed consent was obtained from each subject and all studies were performed according to the tenets of the Declaration of Helsinki.
Genotyping of COL8A2 gene
DNA was extracted from peripheral blood samples by using the QIAamp DNA blood kit (Qiagen, Valencia, USA). All PCR reactions were carried out with 25 ng of genomic DNA as a template in a mixture of PCR buffer, 2.5 mM MgCl2, 200 nM dNTPs, 0.4 pmol of each primer, and 0.75 unit of h-Taq polymerase (Solgent, Daejeon, Korea).8 The genotypes of SNPs spanning the coding region of the COL8A2 gene were determined by single-strand conformational polymorphism (SSCP) analysis and were confirmed by direct sequencing. For SSCP, 10 μl of PCR product were mixed with five volumes of denaturing buffer (90% formamide, 25 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol) and incubated for 5 min at 95°C. Samples were immediately put on ice and loaded on a non-denaturing polyacrylamide gel. After electrophoresis, the DNA bands were visualized by the silver staining method and then those showing variation were directly sequenced. For DNA sequencing, amplified DNA was purified using the QIAquick PCR purification kit (Qiagen) and sequenced directly according to the protocols accompanying the BigDye Terminator cycle sequencing kit (Applied Biosystems, Foster City, USA). The novel mutation, Q455V, was confirmed by analysis of the Fnu4HI RFLP (New England Biolabs, Beverly, MA, USA, Figure 1f). Nucleotide sequences were blasted against the published cDNA sequence of the COL8A2 gene.
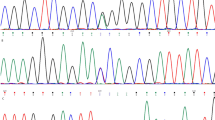
COL8A2 gene mutation screening. Top: Electropherograms showing mutations by direct sequencing; Bottom: Partial DNA sequences of mutation region. (a) Mutation distribution in COL8A2 gene. (b) A35A (c.112G>A) polymorphism in nonhelical region 2 domain of COL8A2, (c) R155Q (c.471G>A) polymorphism in the triple-helical domain of COL8A2, (d) detection of Q455V (c.1370-1371CA>GT) pathogenic mutation in triple-helical region, and (e) detection of G495G (c.1492G>A) and T502M (c.1512C>T) polymorphisms. (f) The novel mutation, Q455V (CA>GT transition), was confirmed by analysis of an Fnu4HI RFLP. The *CA allele was digested into two fragments (308 and 204 bp), whereas the *GT allele was not digested by Fnu4HI (512 bp). Lane M, PCR marker; Lanes 1 and 5, CA/GT heterozygote; Lanes 2, 3, 4, and 6, CA homozygote.
Prediction of the functional effect of amino acid substitutions
Two sequence homology-based programmes were used to predict the likelihood that an amino-acid substitution would have a phenotypic effect: PolyPhen (Polymorphism Phenotyping, http://genetics.bwh.harvard.edu/pph/) and SIFT (Sorting Intolerant from Tolerance, http://blocks.fhcrc.org/sift/SIFT.html).18, 19, 20 SIFT is a sequence homology-based tool that presumes that important amino acids are conserved in the protein family. Hence, changes occurring at well-conserved positions are generally predicted to be deleterious. Substitutions at each position showing normalized probabilities less than a chosen cutoff value are predicted to be deleterious, and those greater than or equal to the cutoff value are predicted to be tolerated. The cutoff value in the SIFT program is a tolerance index of >0.05. The higher the tolerance index, the less functional impact a particular amino acid substitution is likely to have. The level of structural alteration caused by three nonsynonymous mutations was determined by applying the PolyPhen programme, which structurally analyses the amino-acid polymorphism and is used to predict whether an amino acid change is likely to be deleterious to protein function. The prediction is based on the position-specific independent counts (PSIC) score derived from multiple sequence alignments of observations. PolyPhen scores of >2.0 indicate probably damaging to protein function, scores of 1.5–2.0 as possibly damaging, and scores of <1.5 as benign and unknown.18, 20
Statistical methods
Data were analysed using online statistical analysis tools. Statistical differences in allele and genotype frequencies between the case and the control groups were determined using Pearson's chi-squared test or by Fisher's exact test, respectively. The Hardy–Weinberg equilibrium (HWE) of SNPs in patients and controls was calculated using the GenePop programme. The strength of the association was estimated by odds ratio of risk (OR) and 95% confidence intervals (CI) (JavaStat, http://members.aol.com/johnp71/ctab2x2.html).
Results
A total of 25 FECD patients were screened, including 15 patients from six pedigrees and an additional 10 unrelated FECD patients. All FECD patients, including 15 patients from six pedigrees and 10 unrelated patients, ranged in age from 20 to 81, with an average age of 47 years. Among the affected probands from FECD families, diagnosis was made between 21 and 40 years. Sixteen of the patients with FECD were women and nine patients were men. The 73 unrelated control populations aged between 30 and 75 years. PCR—SSCP and direct sequencing were used to screen genetic variations in COL8A2. Through this screening method, we identified three heterozygous missense mutations and two silent mutations: c.112G>A (A35A), c.471G>A (R155Q), c. 1370_1371CA>GT (Q455V), c.1492G>A (G495G), and c.1512C>T (T502M). These mutations are positioned within the COL8A2 triple-helical domain with the exception of mutation A35A (Figure 1a). Among these mutations, the heterozygous two base pair transition of c.1370-1371CA>GT within the triple-helical domain of exon 2, which results in a substitution of valine by glutamine (Q455V), was present in all 12 patients from six pedigrees with a family history of FECD, but only in 2 of 9 unrelated FECD patients (22.2%) (Figures 1d, 1f and 2, Table 1). All FECD patients with Q455V revealed a fine distribution of guttae in contrast to patients with late onset FECD (Figure 3). But this Q455V mutation was not found in unaffected individuals from the same pedigrees or in control subjects (Table 1, Figure 2). The c.112G>A variant within the non-helical region 2 (NC2) of exon 1 (A35A) was also associated with an increased risk for FECD. An increased frequency of the c.112G/G genotype was more prevalent in FECD patients compared to controls (68.8 vs 37.0%, P=0.026, OR=3.75, 95% CI: 1.216–11.460), but the c.112A/A genotype was not found in FECD patient groups. The frequency of the c.112G allele was 0.844 in FECD patients compared to 0.589 in controls (P=0.008), corresponding to an OR for G carriers of 3.77 (95% CI: 1.413–9.984) (Figure 1b, Table 2). The c.1492G/G genotype (G495G) was significantly higher in FECD patients than in the control group (43.8 vs 13.2%, P=0.011, OR=4.90, 95% CI: 1.539–15.759), and the c.1492 G allele frequency was higher in FECD patients compared to controls (0.625 vs 0.388, P=0.018, OR=2.63, 95% CI: 1.210–5.698) (Figure 1e). The two silent mutations were also statistically significant between the affected individuals and the control subjects (Table 2).
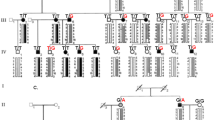
Pedigrees of FECD families (a) FECD-JHK (b) FECD-JYY (c) FECD-PYJ (d) FECD-PJA. Filled symbols, affected individuals; unfilled symbols, unaffected individuals; squares, male; circles, female; arrow, the proband; asterisk, individuals for whom DNA collection and genotyping of COL8A2 was performed. Missense mutations in COL8A2 (mutation order; R155Q, Q455V, and T502M); +, wild type; M, missense mutation.
Although the heterozygous single base pair transitions c.471G>A (R155Q) and c.1512C>T (T502M) were identified in affected individuals, both mutations were also found in control subjects (Table 1). The frequencies of the *G/*G and *G/*A genotypes of c.464G>A (R155Q) were 87.5 and 13.3% in FECD patients and 80.8 and 19.2% in controls, respectively. The *A/*A genotype was not found in both groups. The frequencies of the *C/*C, *C/*T, and *T/*T genotypes of c.1505C>T (T502M) were 75.0, 25.0, and 0.0% in FECD patients and 63.2, 35.5, and 1.3% in controls, respectively. There were no statistically significant differences in the frequencies of R155Q and T502M between the affected individuals and control subjects (Figure 1c and 1e, Table 2). The genotype distributions of all polymorphisms of COL8A2 among the control subjects and the affected individuals were in HWE.
We assessed mutations by two in silico prediction algorithms, PolyPhen and SIFT, to predict the functional impact of amino acid changes (Table 3). The protein sequences of three nonsynonymous mutations were submitted independently to the SIFT programme to determine the tolerance index. All three missense mutations, R155Q, Q455V, and T502M, showed a highly deleterious effect, which was indicated by a tolerance index score of 0.00. The structural levels of alteration were determined by applying the PolyPhen programme, and a PSIC score difference was computed for each mutation. Two missense mutations, Q455V and T502M, were identified as being possibly damaging with a PSIC score of 1.538 and 1.693, respectively; thus, these mutations might significantly affect protein structure. R155Q was predicted to be benign with a PSIC score of 0.900. On the basis of these analyses using SIFT and PolyPhen, Q455V and T502M were predicted to be functionally deleterious.
Discussion
FECD is a primary degenerative disease that accounts for approximately 10% of all corneal grafts.21 FECD is characterized by a pleomorphic, attenuated corneal endothelium with an irregularly thickened Descemet's membrane and central corneal guttatae, which are refractile excrescences prominent on the posterior corneal surface. The endothelial cell abnormality in FECD remains poorly understood. However, on the basis of morphological abnormality, the altered synthesis or assembly of extracellular protein has been suggested to play an important role in the pathogenesis of FECD.
Collagen type VIII α2 (COL8A2) is a major constituent in Descemet's membrane, in which it creates a matrix underneath the endothelial cells, in the form of a hexagonal lattice structure. It is characterized by a short triple-helical domain flanked by one triple-helical domain and two non-triple helical domains at the amino terminus (NC2) and the carboxyl terminus (NC1). In particular, it was reported that mutations in the triple-helical domain of the COL8A2 gene were identified as pathogenic mutations in an FECD-affected family and that these mutations lead to structural alteration in the Descemet's membrane.6, 8, 13, 14 Biswas et al (2001)8 reported that Q455K was detected in both early and late onset familial FECD in individuals originating from Northern England and Australian kindred. Gottsch et al (2005)6 reported that COL8A2 L450W demonstrated segregation with the disease phenotype in American familial FECD with early onset subtype and that it involved massive accumulation and abnormal assembly of collagen VIII within Descemet's membrane.13, 14 However, neither mutation was identified in unrelated FECD patients with typical late onset. Moreover, Biswas et al (2001)8 and Gottsch et al (2005a)6 reported that Q455K and L450W were also associated with the diagnosis of sporadic PPCD, having some features similar to FECD.
In English FECD patients, three missense mutations of the COL8A2 gene, R155Q, R304Q, and R434H, were identified in the common, late onset form of FECD.8 However, Kobayashi et al (2004) concluded that the R155Q and T502M mutations in the COL8A2 gene may not be causative for FECD because heterozygosity was observed in R155Q and T502M mutations not only in Japanese FECD patients but also in control subjects.12 Moreover, a patient with the R155Q and T502M mutations showed no corneal guttae or endothelial cell decrease. Although Aldave et al (2006)15 identified the R434H mutation of the COL8A2 gene in American FECD patients, R434H was also detected in an individual with keratoconus.
In this study, we identified three heterozygous missense mutations and two silent mutations: c.112 G>A (A35A), c.471 G>A (R155Q), c. 1370_1371 CA>GT (Q455V), c.1492 G>A (G495G), and c.1512 C>T (T502M). Among these mutations, the heterozygous two base pair transition of c.1370-1371 CG>AT within the triple-helical domain of exon 2 results in a substitution of valine by glutamine (Q455V). Q455V was present in all patients from six pedigrees of probands and two unrelated FECD patients with FECD early onset subtype, whereas it was not found in unaffected individuals from the same pedigrees or from control subjects or from any of the corneal diseases (Table 1, Figure 2). A slit-lamp examination of patients with Q455V revealed features characteristic of FECD early onset subtype (Figure 3). Thus, the Q455V mutation was presumed to play a pathogenic role in Korean FECD early onset subtype.
Four other previously described polymorphisms in the COL8A2 gene were also detected in the Korean FECD patients. Two silent mutations, A35A and G495G, were also associated with an increased risk for both familial and unrelated FECD patients, but these mutations may be considered nonpathogenic variants because A35A and G495G do not produce a change in the encoded amino acid. Both R155Q and T502M mutations were identified in Korean FECD patients and also in control subjects, but no statistically significant difference for these mutations was observed between Korean FECD patients and normal controls. However, L450W or Q455K mutations, which are known as pathogenic mutations in early onset subtype of FECD, were not found in the Korean FECD probands and unrelated patients of this study.
To predict the potential effects of missense mutation on protein function, we obtained estimates of the impact of three missense mutations through the use of two sequence homology-based programmes, SIFT and PolyPhen. Mutations within highly conserved regions are likely to be of greater phenotypic significance than those within more diverged regions.18, 19 Moreover, several studies indicate that analysing nonsynonymous mutations at the structural level is very important in understanding the functional activity of the protein of interest.22, 23, 24, 25 The SIFT programme was applied to prioritize three missense mutations in the exon 2 coding region of COL8A2. The tolerance index score for each mutation was 0.00, predicting a deleterious effect for each. The structural levels of alteration were then determined by applying the PolyPhen programme in which a PSCI score difference of 1.1 and above is predictive of a structure damaging mutation.18 Two missense mutations, Q455V and T502M, were predicted to be possibly damaging to protein structure with PSIC scores of 1.538 and 1.693, respectively. R155Q was predicted to be a mutation with a benign effect, indicated by a PSIC score of 0.900. Thus, two missense mutations, Q455V and T502M, were identified as potentially important in the identification of FECD caused by the COL8A2 gene. However, we suggest that the mutations associated with Q455V would be of prime importance in the identification of FECD caused by the COL8A2 variants, because the T502M polymorphism showed no statistically significant difference between Korean FECD patients and normal controls.
In this study, we identified a novel mutation, c.1370-1371CA>GT (Q455V), in COL8A2, which was associated with Korean patients with FECD early onset subtype. A Q455V, which is positioned within a short, triple-helical domain of COL8A2, represents a change in the chemical property of the amino acid. This change in chemical property from a nonpolar valine to a polar glutamine is thought to disrupt COL8A2 interaction with two COL8A1 proteins in the hexagonal lattice structure of Descemet's membrane in the posterior collagenous zone.6, 8, 13, 26, 27, 28 Consequently, our data indicate that a Q455V mutation may act as a potential susceptibility mutation, which seem to be associated with FECD predisposition in Korean patients with FECD early onset subtype.
References
Wilson SE, Bourne WM . Fuchs' dystrophy. Cornea 1988; 7: 2–18.
Klintworth GK . The molecular genetics of the corneal dystrophies; current status. Front Biosci 2003; 8: 687–713.
Chiou AG, Kaufman SC, Beuerman RW, Ohta T, Soliman H, Kaufman HE . Confocal microscopy in cornea guttata and Fuchs' endothelial dystrophy. Br J Ophthalmol 1999; 83: 185–189.
Li QJ, Ashraf MF, Shen DF, Green WR, Stark WJ, Chan CC et al. The role of apoptosis in the pathogenesis of Fuchs endothelial dystrophy of the cornea. Arch Ophthalmol 2001; 119: 1597–1604.
Mohamadi P, McDonnell JM, Irvine JA, McDonnell PJ, Rao N, Smith RE . Changing indications for penetrating keratoplasty, 1984–1988. Am J Ophthalmol 1989; 15: 550–552.
Gottsch JD, Sundin OH, Liu SH, Jun AS, Broman KW, Stark WJ et al. Inheritance of a novel COL8A2 mutation defines a distinct early-onset subtype of fuchs corneal dystrophy. Invest Ophthalmol Vis Sci 2005; 46: 1934–1939.
Krachmer JH, Purcell Jr JJ, Young CW, Bucher KD . Corneal endothelial dystrophy; a study of 64 families. Arch Ophthalmol 1978; 96: 2036–2039.
Biswas S, Munier FL, Yardley J, Hart-Holden N, Perveen R, Cousin P et al. Missense mutations in COL8A2, the gene encoding the alpha-2 chain of type VIII collagen, cause two forms of corneal endothelial dystrophy. Hum Mol Genet 2001; 10: 2415–2423.
Rosenblum P, Stark WJ, Maumenee IH, Hirst LW, Maumenee AE . Hereditary Fuchs' dystrophy. Am J Ophthalmol 1980; 90: 455–462.
Sundin OH, Broman KW, Chang HH, Vito EC, Stark WJ, Gottsch JD . A common locus for late-onset Fuchs corneal dystrophy maps to 18q21.2-q21.32. Invest Ophthalmol Vis Sci 2006; 47: 3919–3926.
Sundin OH, Jun AS, Broman KW, Liu SH, Sheehan SE, Vito EC et al. Linkage of late-onset Fuchs corneal dystrophy to a novel locus at 13pTel-13q12.13. Invest Ophthalmol Vis Sci 2006; 47: 140–145.
Kobayashi A, Fujiki K, Murakami A, Kato T, Chen LZ, Onoe H et al. Analysis of COL8A2 mutation in Japanese patients with Fuchs' endothelial dystrophy and posterior polymorphous dystrophy. Jpn J Ophthal 2004; 48: 195–198.
Gottsch JD, Zhang C, Sundin OH, Bell WR, Stark WJ, Green WR . Fuchs corneal dystrophy; aberrant collagen distribution in an L450W mutant of the COL8A2 gene. Invest Ophthalmol Vis Sci 2005; 46: 4504–4511.
Zhang C, Bell WR, Sundin OH, De La Cruz Z, Stark WJ, Green WR et al. Immunohistochemistry and electron microscopy of early-onset fuchs corneal dystrophy in three cases with the same L450W COL8A2 mutation. Trans Am Ophthalmol Soc 2006; 104: 85–97.
Aldave AJ, Rayner SA, Salem AK, Yoo GL, Kim BT, Saeedian M et al. No pathogenic mutations identified in the COL8A1 and COL8A2 genes in familial Fuchs corneal dystrophy. Invest Ophthalmol Vis Sci 2006; 47: 3787–3790.
Aldave AJ, Bourla N, Yellore VS, Rayner SA, Khan MA, Salem AK et al. Keratoconus is not associated with mutations in COL8A1 and COL8A2. Cornea 2007; 26: 963–965.
Yellore VS, Rayner SA, Emmert-Buck L, Tabin GC, Raber I, Hannush SB et al. No pathogenic mutations identified in the COL8A2 gene or four positional candidate genes in patients with posterior polymorphous corneal dystrophy. Invest Ophthalmol Vis Sci 2005; 46: 1599–1603.
Ramensky V, Bork P, Sunyaev S . Human non-synonymous SNPs; server and survey. Nucleic Acids Res 2002; 30: 3894–3900.
Sunyaev S, Ramensky V, Koch I, Lathe III W, Kondrashov AS, Bork P . Prediction of deleterious human alleles. Hum Mol Genet 2001; 10: 591–597.
Ng PC, Henikoff S . SIFT; predicting amino acid changes that affect protein function. Nucleic Acids Res 2003; 31: 3812–3814.
Olsen RJ, Waltman SR, Mattingly TP, Kaufman H . Visual results after penetrating keratoplasty for aphakic bullous keratopathy and Fuchs' dystrophy. Am J Ophthalmol 1979; 88: 1000–1004.
Sullivan LS, Bowne SJ, Birch DG, Hughbanks-Wheaton D, Heckenlively JR, Lewis RA et al. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa; a screen of known genes in 200 families. Invest Ophthalmol Vis Sci 2006; 47: 3052–3064.
Rajasekaran R, Sudandiradoss C, Doss CGR, Sethumadhavan R . Identification and in silico analysis of functional SNPs of the BRAC1 gene. Genomics 2007; 90: 447–452.
Shen J, Deininger PL, Zhao H . Applications of computational algorithm tools to identify functional SNPs in cytokine genes. Cytokine 2006; 35: 62–66.
Bigler J, Sibert JG, Poole EM, Carlson CS, Potter JD, Ulrich CM . Polymorphisms predicted to alter function in prostaglandin E2 synthase and prostaglandin E2 receptors. Pharmacogenet and Genomics 2007; 17: 221–227.
Hopfer U, Fukai N, Hopfer H, Wolf G, Joyce N, Li E et al. Targeted disruption of Col8a1 and COL8A2 genes in mice leads to anterior segment abnormalities in the eye. FASEB J 2005; 19: 1232–1244.
Muragaki Y, Jacenko O, Apte S, Mattei MG, Ninomiya Y, Olsen BR . The alpha 2 (VIII) collagen gene. A novel member of the short chain collagen family located on the human chromosome 1. J Biol Chem 1991; 266: 7721–7727.
Xiong H, Buckwalter BL, Shieh H, Hecht MH . Periodicity of polar and nonpolar amino acids is the major determinant of secondary structure in self-assembling oligomeric peptides. PNAS 1995; 92: 6349–6353.
Acknowledgements
We thank the patients who participated in the study. We also thank SJ Paik, HJ Kim, SH Kim, and JM Seon for excellent technical assistance. We thank the Korea Eye Tissue and Gene Bank, The Catholic University of Korea, and the staff of the Department of Ophthalmology, St Mary's Hospital.This study was supported by the Korea Research Foundation Grant funded by the Korean Government (MOEHRD)(KRF-2006-E00016).
Author information
Authors and Affiliations
Corresponding author
Additional information
This paper was presented in part as a poster at 2007 ARVO (Ft Lauderdale, FL, USA; 7 May 2007) and 2007 Asia ARVO (Singapore; 5 March 2007)
Rights and permissions
About this article
Cite this article
Mok, JW., Kim, HS. & Joo, CK. Q455V mutation in COL8A2 is associated with Fuchs' corneal dystrophy in Korean patients. Eye 23, 895–903 (2009). https://doi.org/10.1038/eye.2008.116
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/eye.2008.116
Keywords
This article is cited by
-
DNA methylation changes and increased mRNA expression of coagulation proteins, factor V and thrombomodulin in Fuchs endothelial corneal dystrophy
Cellular and Molecular Life Sciences (2023)
-
Genetic mutations and molecular mechanisms of Fuchs endothelial corneal dystrophy
Eye and Vision (2021)
-
Collagen type VIII alpha 2 chain (COL8A2), an important component of the basement membrane of the corneal endothelium, facilitates the malignant development of glioblastoma cells via inducing EMT
Journal of Bioenergetics and Biomembranes (2021)
-
The Molecular Basis of Fuchs’ Endothelial Corneal Dystrophy
Molecular Diagnosis & Therapy (2019)
-
Mutations in the TMCO3 Gene are Associated with Cornea Guttata and Anterior Polar Cataract
Scientific Reports (2016)