Abstract
CCCTC-binding factor (CTCF) is a highly conserved zinc finger protein and is best known as a transcription factor. It can function as a transcriptional activator, a repressor or an insulator protein, blocking the communication between enhancers and promoters. CTCF can also recruit other transcription factors while bound to chromatin domain boundaries. The three-dimensional organization of the eukaryotic genome dictates its function, and CTCF serves as one of the core architectural proteins that help establish this organization. The mapping of CTCF-binding sites in diverse species has revealed that the genome is covered with CTCF-binding sites. Here we briefly describe the diverse roles of CTCF that contribute to genome organization and gene expression.
Similar content being viewed by others
Introduction
CCCTC-binding factor (CTCF) is an 82-kDa protein with 11 zinc fingers.1 It was first identified as a transcriptional repressor of the chicken c -myc gene, a regulatory gene that encodes the c-myc transcription factor.2 CTCF is ubiquitously expressed and is highly conserved in eukaryotes.1, 3 CTCF consists of three separate domains: an N-terminal domain, a C-terminal domain and a central domain region with 11 zinc fingers.4 CTCF uses these zinc fingers cooperatively to bind the genome, and the zinc finger domain is especially highly conserved, highlighting its importance in CTCF function.1 All three domains are subject to distinct post-translational modifications.5 CTCF binds between 55 000 and 65 000 sites in mammalian genomes,6 and of these sites, ~50% are intergenic, whereas 35% are intragenic and the rest are promoter proximal,7 showing that CTCF can arrange chromosomal architecture by binding to various sites. CTCF also binds the nuclear matrix and stabilizes nuclear architecture.8 One of exceptional role of CTCF involves its insulator function. Insulators are short nucleotide sequences that set boundaries between nearby genomic domains.9 When CTCF binds to an insulator sequence, the conversation between an enhancer and a gene promoter is impeded and transcription of the gene is blocked.10 In studying gene regulation, it is important to consider regulation at both the locus and genomic levels because gene activity is largely influenced by the spatial positioning of the genome and the stability of genomic architecture.11 This idea emphasizes the importance of CTCF’s ability to bind to a wide range of sequences and control gene expression via the activation or repression of promoters, the insulation of enhancers and the regulation of distant chromatin interactions.3
Role of CTCF as an insulator protein
As briefly mentioned above, an insulator is a DNA sequence that can block the actions of cis-acting elements, such as enhancers, and prevent gene activation.12 Enhancer-mediated activation is a core mechanism of gene regulation in eukaryotes, and enhancers can activate transcription upon activator binding, even when they are positioned far upstream or downstream of the promoter.12, 13 There are two individual loci that were important in the discovery of CTCF’s functions: β-globin and the imprinted H19-Igf2 loci. After discovering that the chicken β-globin locus can block the enhancer activity,14 Bell et al. identified a 42-bp fragment of the chicken β-globin locus that is responsible for the enhancer-blocking activity. They showed that this sequence is the binding site for CTCF and that CTCF has a role in the insulator activity. Through chromosome conformation capture technology, CTCF was shown to help form chromatin loops that encompass elements such as the β-globin gene and the locus control region.15 As these β-globin sites are repositioned through this chromatin loop formation in a way that blocks the enhancer signals, gene transcription is repressed.
Another important locus is the H19/Igf2 locus. The H19 and Igf2 genes are separated from each other by the imprinting control region (ICR), which can be conditionally methylated.16 Methylation of the ICR is a decisive factor in CTCF binding because CTCF only binds the unmethylated ICR on the maternal chromosome. This CTCF binding prevents the communication between the H19-proximal enhancer and the Igf2 promoter, and consequently, Igf2 remains inactivated. Conversely, CTCF cannot bind the paternal ICR because it is methylated,17 and thus, the enhancer is able to activate Igf2 transcription from the paternal chromosome (Figure 1). Upon CTCF binding, various chromatin loops encompassing specific alleles with enhancers and promoters can be formed at the maternal ICR.18 This locus illustrates the basis of how CTCF contributes to insulator activity. Taken together, studies of these two loci showed how CTCF serves as a position-dependent insulator element to block inappropriate enhancer signals and protect against spurious gene activation. This directional enhancer-blocking activity by CTCF seems to be functionally conserved, as the CTCF-binding sites in the insulator region are found in diverse vertebrate species.19 Table 1 shows a list of example sequences that have been tested for insulator function.
H19 and Igf2 gene expression is controlled by CTCF binding at the ICR. CTCF fails to bind to the methylated paternal ICR; thus, the enhancer (E) can induce Igf2 gene expression. However, CTCF binding to the unmethylated maternal ICR acts as a barricade between the Igf2 gene and the enhancer, activating H19 gene expression. CTCF, CCCTC-binding factor; ICR, imprinting control region.
CTCF binding and remodeling of the three-dimensional structure of the genome
Accumulating evidence shows that CTCF aids long-range chromosomal interactions via looping. For example, using chromosome conformation capture and fluorescence in situ hybridization, Hoffman and colleagues have shown that CTCF helps form interchromosomal interactions that co-localize the Igf2/H19 locus on chromosome 7 with the Wsb/Nf1 locus on chromosome 11.20 Deletion of CTCF led to loss of this interchromosomal association and, consequently, changes in Wsb1/Nf1 gene expression.20 This finding provides an example of CTCF’s role in the regulation of chromatin structure and gene expression. CTCF also helps chromatin attach to the nuclear matrix and form functionally distinct regions called topological domains.21 Using ChIP, CTCF was shown to bind numerous genomic sites,22 many of which are conserved among different cell types.23 CTCF is known to bind a CpG-rich consensus sequence that is usually unmethylated, as CTCF preferentially binds to unmethylated elements, such as the H19-Igf2 locus.23 CTCF-binding sites are located at both active and inactive domain boundaries,24 and some are also located at the borders of the lamina-associated domains, where transcriptional activity is low. Steensel and colleagues have created a high-resolution interaction map of the human genome showing that the nuclear lamina interacts with specific genomic regions and organizes the chromosomes into distinct domains.25 They found that CTCF binding is enriched at the lamina-associated domain boundaries, suggesting that CTCF has a role in shaping the three-dimensional chromatin organization. Overall, the characteristics of the identified binding sites influence CTCF’s conformation, its interaction with other proteins and ultimately, genome regulation.
As chromatin interactions are a crucial part of transcriptional regulation, Chromatin Interaction Analysis with Paired-End Tag sequencing (ChIA-PET) is a developing technology that is very useful for studying CTCF’s functions. ChIA-PET allows the analysis of long-range chromatin interactions made by a specific protein.26 In other words, ChIA-PET enables the discovery of interactions between DNA and DNA-associated proteins.27 For CTCF, ChIA-PET revealed ~1500 intrachromosomal and 300 interchromosomal interactions.28 Handoko et al.28 were the first to present a high-resolution CTCF-mediated chromatin interactome map by applying ChIA-PET sequencing in mouse embryonic stem cells. Their data showed that CTCF can create local clusters of genes, direct communication between promoters and regulatory elements, and define the boundaries of chromatin compartments, such as the nuclear lamina. Because CTCF was confirmed to make both intra- and inter-chromosomal interactions,20 it is considered a key organizer that works on the genome at a global scale. A number of studies have revealed that sequences proximal to CTCF-binding sites interact more often with sequences on the same chromosome than do CTCF-distal sites.29 Additional evidence for CTCF as a genome organizer is provided by the observation that its binding sites are enriched with housekeeping genes at the boundaries of topological domains. Ren and colleagues first identified and named the ‘topological domains,’ which are stably inherited in mammalian genomes and mainly act as barriers to the spread of heterochromatin.21 At these domains, CTCF and the cohesin complex hold long-range interactions together and form chromatin loops that define the topological domains. Hadjur and colleagues found that in post-mitotic nuclei without properly working cohesin and CTCF, topological domains become loosened.30 This result shows that CTCF cooperates with other proteins as an organizer and determines which sequences are brought together and what other binding proteins are recruited. Thus, the roles of CTCF extend beyond its original roles as a transcriptional regulator and an insulator; this multivalent protein also orchestrates interactions between distal sequences by binding to chromatin and creating geometrical loops.15
CTCF and the cohesin complex in transcriptional regulation
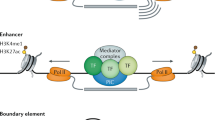
A number of studies have demonstrated the co-localization of CTCF and cohesin on chromosomes, suggesting their functional cooperation. Cohesin is a ring-shaped complex comprised of the subunits SMC1, SMC3 and SCC3.31 It is known to stabilize chromatin loops between enhancers and promoters and also to promote the binding of transcription factors at enhancers.32, 33 Cohesin does not directly bind to DNA, but instead associates with CTCF through its subunit SCC3 at a specific loci.34 In fact, CTCF leads cohesin to its binding sites, and cohesin is required for CTCF to carry out its insulator function.35 Therefore, sequence-specific cohesin binding is dependent on the presence of CTCF, whereas CTCF is not dependent on cohesin for its function.36 These results support the view that CTCF and cohesin work together to assist long-range interactions (Figure 2). CTCF and the cohesin complex maintain higher-order chromatin structures by co-localizing at many sites across the genome. One example is the CFTR locus, which has CTCF-binding insulator sequences. The CFTR locus is activated by enhancers that reach the active promoter through looping. When CTCF or RAD21, a component of the cohesin complex, was knocked down by siRNA, the chromatin structure of the CFTR locus was disturbed, leading to increased gene expression and alterations in histone modifications.37 CTCF also regulates activation of the APP gene, which has highly conserved promoter sequences that share many transcription factor-binding sites. CTCF acts as a transcription factor here that binds the GC-rich -93/-82 promoter region (APBβ) and activates transcription.38
CTCF and the cohesin complex can lead to transcriptional activation or repression in a binding site-dependent manner. For example, when CTCF and cohesin bind to their binding sites and create a chromatin loop that encompasses the enhancer and promoter, transcriptional activation occurs. Conversely, if CTCF and cohesin form a chromatin loop that prevents the enhancer from reaching the promoter, gene expression is repressed. CTCF, CCCTC-binding factor.
Regulation of CTCF activity
As observed for the interaction between CTCF and cohesin, CTCF’s functions are highly affected by its DNA-binding partners. Through the whole genome analyses of CTCF-binding sites, many co-localized proteins were identified, including YY1, Oct4, RNA polymerases and TR.39, 40 Table 2 includes a list of some CTCF-associated proteins and their descriptions. Ying Yang 1 (YY1) is a ubiquitous transcription factor with four zinc fingers that was found to bind along with CTCF to the Trix region of the X chromosome. Renkawitz and colleagues reports that CTCF forms a functional complex with YY1-Oct4-Trix to control the X chromosome. CTCF associated with RNA polymerase II carries out transcription activation, CTCF associated with USF and RNA polymerase I carries out rDNA spacer transcription and CTCF associated with TR causes hormone-sensitive enhancer blocking.39
Moreover, CTCF activity, especially its DNA-binding activity, can be altered by various factors, including DNA methylation. As we have observed in the H19/Igf2 ICR example, methylation is a critical factor that dictates whether CTCF binds at a given locus. Furthermore, Pendone and colleagues were the first to show that only 4 out of the 11 CTCF zinc fingers are essential for strong DNA-binding activity and that each zinc finger differentially contributes to the interaction with a specific locus.41 They also found that cytosine methylation inhibits the DNA binding of zinc finger 7, thus significantly weakening CTCF’s binding affinity.41 At CTCF-binding elements lacking a CpG, CTCF binding was found to be affected by nucleosome repositioning. Lefevre et al.42 showed that proinflammatory stimuli, such as lipopolysaccharide treatment, induces lysozyme gene transcription, which causes nucleosome remodeling. This remodeling eventually leads to the removal of CTCF and its partner cohesin, reversing CTCF-induced gene repression.42 As described, many neighboring factors help CTCF execute its diverse functions by coordinating its zinc fingers to bind to various sequences, and CTCF-binding activity is also regulated by various factors.
Discussion
CTCF is a highly conserved zinc finger protein that performs various regulatory roles in the cell. In this review, we described CTCF’s roles as an insulator protein, a chromatin remodeler and a transcription factor. In addition, we discussed the association of CTCF with cohesin, which is a protein complex that co-localizes with CTCF on the genome. Another primary function of CTCF is as an architectural protein. As the eukaryotic genome is packaged through several levels of organization, its three-dimensional organization is closely related to how genes are expressed. CTCF changes the higher-order chromatin structure and controls the distance between associating domains within and among chromosomes. CTCF is exceptional in that it executes the master role of controlling gene expression. CTCF organizes the genome structure in ways that alter topological domain interactions and ultimately regulate gene expression. Since the first discovery of its role as a transcriptional repressor, many additional studies have revealed CTCF’s various roles and binding sites across the genome. However, many questions still remain about the exact mechanisms by which CTCF carries out its roles, and more information about this multifaceted protein remains to be uncovered.
References
Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G et al. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol 1996; 16: 2802–2813.
Klenova EM, Nicolas RH, Paterson HF, Carne AF, Heath CM, Goodwin GH et al. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol 1993; 13: 7612–7624.
Phillips JE, Corces VG . CTCF: master weaver of the genome. Cell 2009; 137: 1194–1211.
Vostrov AA, Taheny MJ, Quitschke WW . A region to the N-terminal side of the CTCF zinc finger domain is essential for activating transcription from the amyloid precursor protein promoter. J Biol Chem 2002; 277: 1619–1627.
MacPherson MJ, Beatty LG, Zhou W, Du M, Sadowski PD . The CTCF insulator protein is posttranslationally modified by SUMO. Mol Cell Biol 2009; 29: 714–725.
Chen H, Tian Y, Shu W, Bo X, Wang S . Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS One 2012; 7: e41374.
Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008; 133: 1106–1117.
Dunn KL, Zhao H, Davie JR . The insulator binding protein CTCF associates with the nuclear matrix. Exp Cell Res 2003; 288: 218–223.
Gaszner M, Felsenfeld G . Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 2006; 7: 703–713.
Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G . CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell 2004; 13: 291–298.
Misteli T . Spatial positioning. Cell 2004; 119: 153–156.
Bell AC, West AG, Felsenfeld G . The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 1999; 98: 387–396.
Herold M, Bartkuhn M, Renkawitz R . CTCF: insights into insulator function during development. Development 2012; 139: 1045–1057.
Chung JH, Whiteley M, Felsenfeld G . A 5' element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 1993; 74: 505–514.
Splinter E, Heath H, Kooren J, Palstra R-J, Klous P, Grosveld F et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev 2006; 20: 2349–2354.
Pidsley R, Fernandes C, Viana J, Paya-Cano JL, Liu L, Smith RG et al. DNA methylation at the Igf2/H19 imprinting control region is associated with cerebellum mass in outbred mice. Mol Brain 2012; 5: 42.
Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM . CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 2000; 405: 486–489.
Yoon YS, Jeong S, Rong Q, Park K-Y, Chung JH, Pfeifer K . Analysis of the H19ICR insulator. Mol Cell Biol 2007; 27: 3499–3510.
Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep 2005; 6: 165–170.
Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 2006; 312: 269–272.
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012; 485: 376–380.
Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S et al. A map of the cis-regulatory sequences in the mouse genome. Nature 2012; 488: 116–120.
Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res 2012; 22: 1680–1688.
Cuddapah S, Jothi R, Schones DE, Roh T-Y, Cui K, Zhao K . Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 2009; 19: 24–32.
Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008; 453: 948–951.
Li G, Cai L, Chang H, Hong P, Zhou Q, Kulakova EV et al. Chromatin Interaction Analysis with Paired-End Tag (ChIA-PET) sequencing technology and application. BMC Genomics 2014; 15 (Suppl 12): S11.
de Laat W, Dekker J . 3C-based technologies to study the shape of the genome. Methods 2012; 58: 189–191.
Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet 2011; 43: 630–638.
Yaffe E, Tanay A . Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat Genet 2011; 43: 1059–1065.
Sofueva S, Yaffe E, Chan W-C, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J 2013; 32: 3119–3129.
Nasmyth K, Haering CH . Cohesin: its roles and mechanisms. Annu Rev Genet 2009; 43: 525–558.
Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 2010; 467: 430–435.
Faure AJ, Schmidt D, Watt S, Schwalie PC, Wilson MD, Xu H et al. Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules. Genome Res 2012; 22: 2163–2175.
Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM . Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 2008; 27: 654–666.
Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 2008; 132: 422–433.
Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 2008; 451: 796–801.
Gosalia N, Neems D, Kerschner JL, Kosak ST, Harris A . Architectural proteins CTCF and cohesin have distinct roles in modulating the higher order structure and expression of the CFTR locus. Nucleic Acids Res 2014; 42: 9612–9622.
Chen X-F, Zhang Y-W, Xu H, Bu G . Transcriptional regulation and its misregulation in Alzheimer’s disease. Mol Brain 2013; 6: 44.
Weth O, Renkawitz R . CTCF function is modulated by neighboring DNA binding factors. Biochem Cell Biol 2011; 89: 459–468.
Zlatanova J, Caiafa P . CTCF and its protein partners: divide and rule? J Cell Sci 2009; 122 (Pt 9): 1275–1284.
Renda M, Baglivo I, Burgess-Beusse B, Esposito S, Fattorusso R, Felsenfeld G et al. Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J Biol Chem 2007; 282: 33336–33345.
Lefevre P, Witham J, Lacroix CE, Cockerill PN, Bonifer C . The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol Cell 2008; 32: 129–139.
Furlan-Magaril M, Rebollar E, Guerrero G, Fernandez A, Molto E, Gonzalez-Buendia E et al. An insulator embedded in the chicken alpha-globin locus regulates chromatin domain configuration and differential gene expression. Nucleic Acids Res 2011; 39: 89–103.
Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG et al. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. PNAS 2002; 99: 6883–6888.
Murai J, Ikegami D, Okamoto M, Yoshikawa H, Tsumaki N . Insulation of the ubiquitous Rxrb promoter from the cartilage-specific adjacent gene, Col11a2. J Biol Chem 2008; 283: 27677–27687.
Chernukhin I, Shamsuddin S, Kang SY, Bergstrom R, Kwon Y-W, Yu W et al. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol 2007; 27: 1631–1648.
Liu Q, Yang B, Xie X, Wei L, Liu W, Yang W et al. Vigilin interacts with CCCTC-binding factor (CTCF) and is involved in CTCF-dependent regulation of the imprinted genes Igf2 and H19. FEBS J 2014; 281: 2713–2725.
Defossez P-A, Kelly KF, Filion GJP, Pérez-Torrado R, Magdinier F, Menoni H et al. The human enhancer blocker CTC-binding factor interacts with the transcription factor Kaiso. J Biol Chem 2005; 280: 43017–43023.
Acknowledgements
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP; National Honor Scientist Program). SK is supported by a BK21 fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Kim, S., Yu, NK. & Kaang, BK. CTCF as a multifunctional protein in genome regulation and gene expression. Exp Mol Med 47, e166 (2015). https://doi.org/10.1038/emm.2015.33
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/emm.2015.33
This article is cited by
-
CDK9 inhibition induces epigenetic reprogramming revealing strategies to circumvent resistance in lymphoma
Molecular Cancer (2023)
-
SHIELD: a platform for high-throughput screening of barrier-type DNA elements in human cells
Nature Communications (2023)
-
Trio-binning of a hinny refines the comparative organization of the horse and donkey X chromosomes and reveals novel species-specific features
Scientific Reports (2023)
-
Atypical Modes of CTCF Binding Facilitate Tissue-Specific and Neuronal Activity-Dependent Gene Expression States
Molecular Neurobiology (2023)
-
HSFA1a modulates plant heat stress responses and alters the 3D chromatin organization of enhancer-promoter interactions
Nature Communications (2023)