Abstract
Peloponnese has been one of the cradles of the Classical European civilization and an important contributor to the ancient European history. It has also been the subject of a controversy about the ancestry of its population. In a theory hotly debated by scholars for over 170 years, the German historian Jacob Philipp Fallmerayer proposed that the medieval Peloponneseans were totally extinguished by Slavic and Avar invaders and replaced by Slavic settlers during the 6th century CE. Here we use 2.5 million single-nucleotide polymorphisms to investigate the genetic structure of Peloponnesean populations in a sample of 241 individuals originating from all districts of the peninsula and to examine predictions of the theory of replacement of the medieval Peloponneseans by Slavs. We find considerable heterogeneity of Peloponnesean populations exemplified by genetically distinct subpopulations and by gene flow gradients within Peloponnese. By principal component analysis (PCA) and ADMIXTURE analysis the Peloponneseans are clearly distinguishable from the populations of the Slavic homeland and are very similar to Sicilians and Italians. Using a novel method of quantitative analysis of ADMIXTURE output we find that the Slavic ancestry of Peloponnesean subpopulations ranges from 0.2 to 14.4%. Subpopulations considered by Fallmerayer to be Slavic tribes or to have Near Eastern origin, have no significant ancestry of either. This study rejects the theory of extinction of medieval Peloponneseans and illustrates how genetics can clarify important aspects of the history of a human population.
Similar content being viewed by others
Introduction
Peloponnese was peopled by a series of migrations that spanned almost nine millennia. Early migrants arrived from Anatolia ca 9000 BCE1, 2 and established in the peninsula several Neolithic sites.2 The Myceneans3 who developed an advanced Bronze Era civilization, either migrated from the north around 2200 BCE4, 5 or were the descendants of the original Neolithic migrants.1 The next migration took place at the beginning of the first millennium BCE with the invasion of Peloponnese by the Dorian Greeks, referred in the Greek tradition as the return of the Heraclids.6 The subsequent 14 centuries were marked by quantitative changes of the population due to wars and epidemics but no qualitative effects from migrations of new population groups. Changes in population structure started in the beginning of the medieval period with the migrations of the Slavs to the Balkans.7, 8 The effects of these migrations have dominated the historiography of Peloponnese during the last 170 years.
In 1830 CE, the German historian Jacob Philipp Fallmerayer presented his theory of disappearance of the Greek nation and its substitution by Slavs.9 Fallmerayer proposed that during the 6th century CE, large armies of Avars and Slavs overran the Balkans and eliminated the populations of the Hellas, who up to that period had successfully survived the attacks of barbarians and the religious suppression by the Byzantines. The Peloponnesean Greeks, except for few remnants enclosed in coastal castles, were slaughtered or forced to leave and Peloponnese was inhabited by Slavic tribes. The Slavs kept their identity for few centuries but eventually they were Hellenized under the influence of the Orthodox Church and interactions with Hellenized Asia Minor populations who were settled in Peloponnese by the Byzantines. Since the time Fallmerayer’s theory was published, a debate on the question of the ancestry of Peloponneseans has raged among historians (reviewed in Curta,10). Of note is that in spite of their diametrically different views, all historians have been using the same medieval written sources.
Controversies are rather common in historiography and result to endless debates among scholars. Controversies concerning the ancestry of populations can potentially be resolved by genetic analysis. In this paper, we use genome-wide data to study the genetic structure of the Peloponnesean populations and compare them with other populations of the world. We observe characteristic patterns of genetic differentiation within Peloponnese, we examine their possible causes and we focus on the question of the impact of Slavic migrations on the genetic structure of the Peloponnesean populations. Our results reject the theory of extinction of medieval Peloponnesean Greeks and their replacement by Slavic and Asia Minor settlers.
Subjects and methods
Design of the study and populations studied
The study has been reviewed by the Institutional Review Board of the University of Washington and the ethical committees of several provisional hospitals. We focused on the rural population. We analyzed a total of 241 samples genotyped with the Illumina Infinium Omni 2.5–8 arrays. This is a novel data set collected under the auspices of our study. Subjects were included in the study if all four grandparents originated from the same village or from villages that were <10 kilometers apart. The ages of most participants ranged between 70 and 90 years (the oldest subject was 107 years old); hence their grandparents were born between 1860 and 1880. In the 1861 census the population of Peloponnese was 578 598 individuals. At that time the economy of Peloponnese was exclusively agricultural and over 85% of the population was living in small villages and hamlets. We sampled all the districts of Peloponnese (Figure 1a and Supplementary Table 1) and also focused on two culturally distinct subpopulations, the Tsacones and the Maniots. To compare the Peloponneseans with other populations we analyzed samples from published data sets and data sets generated by our studies (Supplementary Table 2 and Supplementary Figure 1). Merging genotypes from different sources and quality control were done as described.11
Substructure of the Peloponnesean populations. (a) Map of Peloponnese showing the populations studied. Each dot corresponds to the origin of a participant. (b) PCA illustrating the separation of Peloponneseans in three groups. On the left is placed the population of Tsakones (north: open circles, south: green dots). On the right are placed the populations of Maniots (Deep Mani, East and West Tayetos.). All the remaining Peloponneseans are clustered in the center. (c) PCA analysis without the Maniot and Tsakones populations showing a partial separation of the population of Laconia. (d) IBD analysis illustrating the separation of Peloponneseans in three subpopulations. The color indicates the log10 of the average proportion of genome shared IBD between individuals in two populations, where higher values (less negative; red color) indicates a higher pairwise proportion of genome shared IBD, and lower values (more negative; yellow color) indicates a lower proportion of genome shared IBD. (e) ADMIXTURE analysis. Notice the distinct structure of the Maniots and the Tsakones and their clear cut separation from all other Peloponneseans in all values of K.
Identity by descent
The data used for the identity by descent (IBD) analysis were derived from data sets listed and referenced in Supplementary Table 2. Two individuals share a genomic segment identical by descent if they have inherited the segment intact from a common ancestor, without intervening recombination. Allele strand, reference and alternate alleles were aligned with the 1000 Genomes European populations (CEU, GBR, TSI, FIN and IBS) using the conform-gt utility (http://faculty.washington.edu/browning/conform-gt.html) prior to running Beagle. Marker exclusion criteria were unconfirmed strand orientation, minor allele frequency <1%, Hardy–Weinberg P-value <10−6, and >2% missing data. After applying these criteria, 560 891 SNPs were used for the IBD analysis.
We used the Refined IBD algorithm implemented in Beagle 4.1(ref. 12) to phase the data and infer IBD segments. We set niterations=160 and used default values for all for all other parameters, including requiring a LOD score of 3, which means that the probability of the observed genotype data for a pair of samples in the inferred IBD segment is at least 1000 times greater under an IBD model than under a non-IBD model. Based on the software recommendations and marker density, we set ibdtrim=22. We used the HapMap genetic map to infer genetic length and excluded IBD segments with length <2 cM, since refined IBD has a low false-positive rate when using this threshold.12
R13 was used to generate plots and summarize IBD distributions. The heat map summarizes average pairwise IBD between chromosomes from different individuals in the populations being compared (see footnote 4 in Supplementary Table 3 for details), where higher average pairwise IBD indicates a closer genetic relationship between populations.
Principal Component Analysis (PCA)
We used Eigenstrat14 as well as our own MatLab implementation of PCA.15, 16
Estimating population admixture
We used the ADMIXTURE v1.22 software for all our admixture analyses.17 Cross validation errors of ADMIXTURE analysis ranged between 0.42–0.47 (Figure 1e), 0.55–0.58 (Figure 2c) and 0.56–0.62 (Figure 4d) with smaller error at K=2.
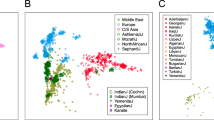
Genetic similarity of Peloponneseans, Sicilians and Italians. PCA analysis of several European populations. (a) Notice the north to south distribution of the populations and that the Peloponneseans are placed to the far right of the graph and overlap with the Sicilians. (b) PCA analysis of Southern European populations illustrating the close relationship between Peloponneseans Sicilians and Italians (TSI is an Italian population) (c) Network analysis illustrating the high connectivity between the Peloponnesean populations as well as between the Peloponneseans, the Sicilians and the Italians. Notice the distance between Peloponneseans and the Slavic, and Near Eastern populations. Peloponneseans are connected with the Near Eastern populations through Crete and Dodecanese.
Testing the hypothesis of Slavic origin of culturally distinct Peloponnesean populations. PCA comparisons of (a) The Maniots of Deep Mani (b) The Maniots of Tayetos (c) The Tsakones, with populations of the Slavic homeland (Ukrainians, Polish, Russians and Belarusians). Notice the broad separation between the Slavs and the Peloponnesean populations. (d) ADMIXTURE analysis shows the complete separation of Maniots and Tsakones from the Slavs in all K values.
Quantitative analysis of ADMIXTURE output
Given a target population X and reference populations Y, Z and so on, we were interested in quantifying the amount of ancestry of population X that is captured by populations Y, Z and so on. Towards that end we devised a new approach to quantitatively analyze the output of ADMIXTURE. Recall that ADMIXTURE, for a particular value of K, will represent each sample using K coordinates. Thus, for a particular value of K and for a particular population Y with n samples, we can represent the output of ADMIXTURE for this population as an n-by-K table. Then, for each reference population Y, we summarize this n-by-K matrix using its top right singular vector only; in all our analyses, the top singular value corresponding to the top right singular vector captured at least 80% of the reference population variance as represented by ADMIXTURE. Let vY be the top right singular vector (a K-dimensional vector) for population Y; similarly, let vZ be the top right singular vector (a K-dimensional vector) for population Z and so on. Now that we have represented the ADMIXTURE output for each population as a K-dimensional signature vector, we can apply standard vector space calculus in order to answer our original question: how much of the ancestry of population X is captured by population Y, or population Z and so on. More specifically, in order to compute the percentage of the ancestry of population X that is captured by population Y, we compute the percentage of the norm of VX that is captured (in projection sense) by vY. Formally, we compute

which returns a value between zero and one. In the above, VX denotes the m-by-K matrix representing the m samples of population X with respect to the K coordinates returned by ADMIXTURE. The notation

indicates the pseudoinverse of the vector vY, which is equal to the transpose of the vector vY, suitably normalized. It is also worth noting that the norm used in the above equation is the standard matrix Frobenius norm. In order to quantify the amount of ancestry of population X that is captured by both populations Y and Z, we form the K-by-2 matrix V=[vY vZ] whose columns are the vectors vY and vZ and we compute

In the above equation, V+ denotes the pseudoinverse of the matrix V; the matrix VV+ is a projector on the subspace spanned by the column space of V. Thus, we basically extract from the matrix VX the part of VX that is captured by the (subspace spanned by the) vectors vY and vZ.
Network analysis
To better visualize and understand the connection between the populations included in our study, we performed a network analysis on the results of ADMIXTURE, using a method presented in reference.11
Results
The substructure of the Peloponnesean populations
On PCA analysis the populations are arranged in the form of an inverted capital letter V (Figure 1b). The left of this formation is occupied by the population of Tsakones who inhabit the east slopes of Mount Parnon and the adjacent costal area (Figure 1a). The right of the formation is occupied by the populations of Maniots who inhabit the east and west slopes of mount Tayetos and the southern area of the promontory, the so called Deep Mani (Figure 1a). All other Peloponneseans cluster in the tip of the letter V (Figures 1b and c). The ADMIXTURE analysis of Figure 1e shows that the Maniots and Tsakones are clearly separated from each other and from all other Peloponnesean populations. Correlations between geographic coordinates and the two principal components (Table 1) are compatible with gradients in gene frequencies from north to south across all Peloponnese, along the Ionian coast, across Arcadia, as well as within Laconia and between the slopes of Tayetos and Deep Mani.
IBD analysis (Figure 1d) confirms the existence of distinct Peloponnesean subpopulations. In the populations of Maniots, individuals share on average 0.25% of their genome (or 35–36 cM) identical by descent, with 95% of pairs of individuals sharing at least one IBD segment (Supplementary Table 3). Similarly, the two Tsakones populations have a very high average pairwise IBD sharing of 0.66% of their genome, or 94 cM; every pair of individuals shares at least one IBD segment. Laconia is fairly closely related to both Deep Mani and to the Tsakones, based on the darker blue for those pairings in the heat map of Figure 1d. The heat map also revealed a larger separation between the southern and northern Peloponnesean populations, with the populations of Arcadia, Messenia, Elis, Achaea, Corinthia and Argolis forming their own subgroup. Corinthia and Argolis are the most strongly related (by IBD) of this subset of populations, sharing nearly 10 cM IBD on average.
Genetic similarity with Sicilians and Italians
As anticipated from the results of previous studies,18, 19, 20 the Peloponneseans are genetically placed very close to the Sicilians and Italians (Figures 2a and b) but they differ from several other populations we compared them (see Supplementary Figure 2). Network analysis (Figure 2c), highlighted the interconnections of Peloponnesean populations as well as the connections between Peloponneseans, Italians and Sicilians; in this network analysis, Sicilians and Italians serve as a bridge between Peloponneseans and other European populations (Basque, Andalusians and French). Slavic populations are placed far away from the Peloponneseans as are the Near Eastern populations. The latter are connected to the Peloponnesus via the islands of Crete and the Dodecanese.
Testing the theory of extinction of the medieval Peloponnesean Greeks
This theory allows a specific prediction about the genetic ancestry of the Peloponneseans: the great majority, if not all, of Peloponnesean ancestry should be Slavic. We compared, the Peloponneseans (except for Maniots and Tsakones) with populations of the Slavic homeland from which the sixth century Slavs should have originated. The exact location of the Slavic homeland is debated7, 8 but it is placed north of Danube,7 between the Oder and Dnieper rivers and includes areas inhabited by Polish, Ukrainian, Russian and Belarusian populations. PCA analysis showed a clear separation of Peloponneseans from the Slavic populations (Figure 3a). By ADMIXTURE analysis (Figure 3c) the Peloponneseans and the Slavic populations form separate clusters with a small degree of gene flow from the Slavic to the Peloponnesean cluster.
Testing the theory of replacement of medieval Peloponesians by Slavs and Asia Minor settlers. (a) PCA analysis shows the broad separation of Peloponneseans from four populations of the Slavic homeland (Ukrainians, Polish, Russians and Belarusians). (b) PCA comparisons of the Peloponneseans with three Greek-speaking Asia Minor populations shows only partial overlap with the population of the Asia Minor Aegean coast. (c) ADMIXTURE analysis illustrates the wide separation of Peloponneseans from the Slavs in all values of K.
Fallmerayer hypothesized that the Hellenization of the Peloponnesean Slavs was accelerated by the transfer to the Peloponnese of Hellenized populations from Asia Minor.9 We tested this hypothesis by comparing the Peloponneseans with three Greek-speaking populations of Asia Minor: a western-coastal population sample extending from the Propontis in the north to Alikarnassos (Bodrum) in the south; a northern population from Pontus, that is, the coast of Black Sea and the Asia Minor interior corresponding to the current northern Turkey; and a central Anatolian population from Cappadocia. All these populations are separated from the Peloponneseans by PCA (Figure 3b). The small degree of overlap between Peloponnese and the population of the Asia Minor coast (Figure 3b) is expected for neighboring Greek populations. The Byzantines frequently moved Armenians to achieve political objectives21 and Fallmerayer proposed that Armenians were included in the populations transferred to Peloponnese by the Byzantines. Peloponneseans differ from the Armenians by PCA and ADMIXTURE analysis (Supplementary Figure 3). Collectively, these results are incompatible with the theory of extinction of the medieval Peloponneseans and their replacement by Slavic and Asia Minor settlers.
The medieval ancestry of the populations of Mani
The Maniots differ from all other Peloponneseans by PCA (Figure 1b) and ADMIXTURE (Figure 1e) analysis. They also differ from mainland, island and Asia Minor Greek populations (data not shown) and from all the other populations of Supplementary Figure 4, which have been compared by PCA analysis, but they partially overlap with the Sicilians and the Italians.
In his treatise on the administration of the Byzantine Empire,22 the Emperor Constantin Porphyrogenitus describes how two Slavic tribes, the Mellingi and the Ezeritae, were forced by the Byzantines to withdraw to the slopes of Tayetos. The Mellingi continued to be mentioned in the Peloponnesean history untill the 15th century.23 Because of the writings of Porphyrogenitus we sampled separately the populations of the East and the West slopes of the Tayetos and the Deep Mani. By PCA (Figure 4a) and ADMIXTURE (Figure 4d) the populations of Tayetos are distinct from the populations of the Slavic homeland. Fallmerayer argued that the inhabitants of Deep Mani are of Slavic origin.9 PCA (Figure 4b) and ADMIXTURE (Figure 4d) analyses makes this hypothesis unlikely.
As an alternative origin of the Maniots, Fallmerayer proposed that they are descendants of Mardaites.9 This medieval warrior tribe used to inhabit the mountainous regions between Asia Minor and Syria but in late seventh century CE was resettled by the Byzantines in Asia Minor and other areas of the Empire.24 The Mardaites have disappeared from history but oral tradition claims that they are the ancestors of the Maronites of Lebanon, although this claim has been disputed.25 PCA and ADMIXTURE analyses failed to show any close relationship between Maniots and the Maronites (Supplementary Figure 5).
The question of Slavic ancestry of Tsakones
The Tsakones of the eastern slopes of Mount Parnon differ from all other Peloponneseans (Figures 1b and e) and from all other populations of Supplementary Figure 6 we have compared them. They used to speak a dialect of Doric origin26 which was not comprehended by the other Peloponneseans. Their name was considered by medieval authors to represent a corruption of the word Lacones (Tsakones=Lacones). Fallmerayer argued against a Doric origin of the Tsakones and, instead, proposed that they were the descendants of a Slavic tribe that had migrated to Peloponnese before the flood of the Slavic settlers reached the peninsula. PCA (Figure 4c) and ADMIXTURE (Figure 4d) analyses argue against this hypothesis.
Quantitative assessment of the ancestry of Peloponneseans
To quantify the findings of the ADMIXTURE analyses, we employed a method for the meta-analysis of the ADMIXTURE output that treats the output as a set of vectors in a K-dimensional space (for a particular value of K between four and eight). Each population is then summarized by a single vector (using PCA) and vector space calculus is used in order to identify the percentage of ancestry of a target population that is captured by one or more reference populations. Notice that our choice to summarize each population by a single vector is akin to computing the mean ADMIXTURE output for a particular population. In most cases, ADMIXTURE returns a homogenous structure for a particular population and thus the top principal component is a good summary of the sample vectors returned by ADMIXTURE.
First we focused on the ADMIXTURE analysis of Figure 3c which includes seven Peloponnesean populations (Argolis, Corinthia, Achaea, Elis, Arcadia, Messenia and Laconia), four Slavic populations (Belarusians, Russians, Polish and Ukrainians), three Southern European populations (Italians, Basque and Andalusians) and the French. The results of Table 2 show that there is considerably more shared ancestry between the Peloponneseans and the French, Andalusians and Italians compared to the shared ancestry between the Peloponneseans and the Slavic populations. The average shared ancestry with French ranges from 39 to 42%; with Andalusians from 53 to 62%; and with the Italians from 85 to 96%. In contrast, the average shared ancestry with the Slavic populations is always <15%. Therefore, the Peloponneseans are genetically much more distinct from the Slavic populations and are much more similar to Southern European populations. We also observe that the Basques, (a population that is well-known to be isolated and genetically different from even its neighboring Iberian populations) are very distinct from all populations in our analysis. This is precisely why we included them in these ADMIXTURE meta-analyses: on average Basques share <4% of common ancestry with any Peloponnesean population. Notice that this number is relatively close to the average ancestry shared between the Peloponnesean populations and the Belarusians, Polish and Ukrainians. All these populations share between 5.2 and 8.5% of common ancestry with the Peloponnesean populations. These Slavic populations are, from a genetic perspective, approximately as far apart from the Peloponneseans as are the Basques.
We next determined the shared ancestry between the five distinct Peloponnesean populations (Deep Mani, West and East Tayetos, North and South Tsakonia), the Slavs, the southern European populations, the French and the Basque. The ADMIXTURE plot of Figure 4d and the data of Table 3 show that the amount of shared ancestry between these five Peloponnesian populations and the Slavic populations is very low. The ancestry Deep Mani shares with Belarusians, Polish and Ukrainians ranges from 0.7 to 1.0%. East and West Tayetos share from 4.9 to 8.6% ancestry with the three Slavic populations which is five to eight times higher than that of Deep Mani but lower to the ancestry the other Peloponnesans share with the Slavs. Slightly lower, compared to the other Peloponneseans, is the ancestry shared between West/East Tayetos and the Russians (8.6–10.9%). The ancestry North and South Tsakonia shares with the Slavs ranges from 4 to 8% and 0.2 to 0.9%, respectively. Compared to the very low ancestry shared with the Slavs, South Tsakonia and Deep Mani share 14 and 25% ancestry with the Italians. North Tsakonia, East and West Tayetos share from 41 to 57% ancestry with the Italians. Again, the Basques are isolated from the five Peloponnesean populations.
Discussion
Our analysis of the genetic ancestry of the Peloponnesean populations and their relationships with the Slavs and other Europeans settles a historical controversy that has persisted for over 170 years. This controversy illustrates the problems historians face in their efforts to reconstruct population history on the basis of inadequate written sources. Fallmerayer based his theory of extinction of the medieval Peloponnesean Greeks on the writings of few early and two middle-medieval Byzantine authors. The early sources were very short comments in texts of 6th and 7th century historians and ecclesiastic authors.9 The middle-medieval documents were a letter by an eleventh century Patriarch of Constantinople and the writings of tenth century Emperor Constantine Porphyrogenitus. Fallmerayer’s theory created sensation among historians. An early rebuttal was published by the Greek historian Paparrigopoulos27 who examined the same sources Fallmerayer have used to construct his theory and reached different conclusions, that is, that the Slavs did not reach the Greek proper during the sixth century and, when they arrived, they did not slaughter the local population. The many historians who have contributed to the very extensive literature on this topic during the last century (partially summarized in Curta10 and Zakythenos28) usually either accept or reject the theory of extinction of the Peloponnesean Greeks. It seems that personal philosophies influence the historians’ judgment. Fallmerayer was an educator and journalist turned historian, a liberal intellectual for his time and a slavophobe who feared the increasing influence of Russia in the Balkans at the expense of the Ottoman Empire. Paparrigopoulos was a Greek historian who was promoting the idea of the continuity of the Greek ethnicity during the medieval period. The findings of our study settle these issues and provide a direct test of the theory of the extinction of the medieval Peloponnesean Greeks. It is clear that the Slavs settled in Peloponnese, as the quantitative measurements of Slavic ancestry indicate (Tables 2 and 3). It also seems that their numbers were relatively small compared to the size of the local population as the low levels of Slavic ancestry of the Peloponnesean populations indicate.
In his book on the Administration of the Empire22 Constantin Porphyrogenitua describes the wars between the Byzantines and two Slavic tribes, who initially had settled the lowland Laconia but were forced to withdraw to the security of the slopes of the mount Tayetos, in order to avoid subjugation to Byzantine rule. Porphyrogenitus tells us that the slopes of Tayetos were Slavic lands; however, our analyses show low levels of Slavic ancestry in the populations of Tayetos. The most reasonable interpretation for the discrepancy between the medieval text and the genetic data is that the size of the Slavic settlements in the slopes of Tayetos was small and the Slavic population was diluted by migrations from Deep Mani during the subsequent centuries. Despite its inhospitable environment, Deep Mani was densely populated29 and there is historical evidence for high mobility and migrations of Maniots.30 A gene flow path from Deep Mani to the slopes of Tayetos is also suggested by our PCA analysis and the correlations between geographic coordinates and principal components.
The striking difference between the Tsakones and the remaining Peloponneseans on PCA and ADMIXTURE analysis can be attributed to isolation by distance. Geographic isolation explains the retention of their dialect. In ancient times the land of Tsakones, then called Cynouria, was inhabited by Doric speaking Ionians6 and because of its geography was most likely isolated from the other Peloponneseans. Isolation by distance is also the likely explanation of the findings in the populations of Mani. Porphyrogenetus in his writing about the Slavs of Tayetos also asks what happened to the ancient inhabitants of Laconia, the Hellenes, who continued to adhere to the ancient Greek religion.22 He finds them withdrawn in the inhospitable, agriculturally poor and rocky area of southern Tayetos, the area which we refer to here as the Deep Mani. Ancient DNA studies could perhaps test whether there is any relationship between Maniots and ancient Lacons or between Tsakones and ancient Ionians.
References
Renfrew C : Archaeology and Language, the Puzzle of Indo-European Origins. London: Jonathan Cape, 1987.
Perles C : The Early Neolithic in Greece. Cambridge: Cambridge University Press, 2001.
Chadwick J : The Mycenean World. Cambridge: Cambridge University Press, 1976.
Hooker JT : The Coming of the Greeks. Claremont: Regina Books, 1999.
Gimbutas M Proto-Indo-European culture: the kurgan culture during the fifth, fourth and third millennia BC. In: Cardona G, Hoenigswalt HM, Senn A (eds): Indo-European and Indo-Europeans. Philadelphia: University of Pennsylvania Press, 1970, pp 155–197.
Herodotus : The Histories The Loeb Classical Library. Cambridge: Harvard University Press, 1981.
Curta F : Southeastern Europe in the Middle Ages 500–1250. Cambridge: Cambridge University Press, 2006.
Fine JVA : The Early Medieval Balkans: A Critical Survey from the Sixth to the Late Twelfth Century. Ann Arbor: The University of Michigan Press, 1991.
Fallmerayer JP Geschichte der halbinsel morea wahrend des mittelalters: ein historischer versuch; Untergang Der Peloponnesischen Hellenen Und Wiederbevolkerung Des Leeren Bodens Durch Slavische Volksstamme. In: JG Cotta (ed): Stuttgart und Tubingen, Germany, 1830, Vol 1..
Curta F : The Edinburgh History of the Greeks, C 500 to 1050. Edinburgh: Edinburgh University Press, 2011.
Paschou P, Drineas P, Yannaki E et al: Maritime route of colonization of Europe. PNAS 2014; 111: 9211–9216.
Browning BL, Browning SR : Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 2013; 194: 459–471.
Core Team R : A Language And Environment For Statistical Computing. Vienna: R Foundation for Statistical Computing, 2014.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D : Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909.
Paschou P, Mahoney M, Pakstis A, Kidd JR, Kidd KK, Drineas P : Inter- and intrapopulation genotype reconstruction from tagging SNPs. Genome Res 2007; 17: 96–107.
Paschou P, Ziv E, Burchard EG et al: PCA-correlated SNPs for structure identification in worldwide human populations. PLoS Genet 2007; 3: 160.
Alexander DH, Novembre J, Lange K : Fast model-based estimation of ancestry in unrelated individuals. Genome Res 2009; 19: 1655–1664.
Cavalli-Sforza LL, Menozzi P, Piazza A : The History and Geography of Human Genes. Princeton: Princeton University Press, 1994.
Di Gaetano C, Cerutti N, Crobu F et al: Differential Greek and northern African migrations to Sicily are supported by genetic evidence from the Y chromosome. Eur J Hum Genet 2009; 17: 91–99.
Fiorito G, Di Gaetano C, Guarrera S et al: The Italian genome reflects the history of Europe and the Mediterranean basin. Eur J Hum Genet 2016; 24: 1056–1062.
Charanis P : The Armenians in the Byzantine Empire. Byzantinislavica 1961; 22: 196–240.
Porphyrogenitus C : De Administrando Imperio. District of Columbia: Dumbarton Oaks, 1967.
Rodd R : The Princes of Achaia and the Chronicles of Morea, A Study of Greece in the middle ages. London: Edward Arnold, 1907.
Charanis P : The Transfer of Population as a Policy in the Byzantine Empire. Comp Stud Soc Hist 1961; 3: 140–154.
Moosa M : The Maronites in History. Syracuse: Syracuse University Press, 1986.
Scutt CA : The Tsakonian Dialect. Ann Brit School at Athens 1913; 19: 133–173.
Paparrigopoulos K : Πɛρί ɛποικίσɛως Σλαβικών τινων ϕυλών ɛις την Πɛλοπόννησον. About The Settlement Of Slavic Tribes in Peloponnese: Athens 1843.
Zakythenos AD Οι Σλαβοι ɛν Ελλαδι (The Slavs in Greece). Aetos: Athens, 1945.
Komis K : Πληθυσμός και οικισμοί της Μάνης 15ος -19ος αιώνας (Settlements and Population of Mani, 15 to 19 century), Ioannina 2005.
Nicholas N : A history of the Greek colony of corsica. J Greek Diaspora 2005; 31: 33–78.
Acknowledgements
This study was supported with funds from the Lucille P Markey Charitable Trust to GS; National Science Foundation grants to PD; Altius Institute for Biomedical Sciences to JS; ARISTEIA II Programme and NSRF 2007–2013 grant to PP; and NIH HG008359 grant to BLB. AP was supported by the NSF Graduate Research Fellowship Program DGE-1256082. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We wish to thank the following physicians and health center personnel for their invaluable help during the field study. Arcadia: P Bourtzoukli, Argolis: D Kostakis, P Kambosos, E Spyropoulou, Achaia: N Razis, A Kouroukli-Symeonidou, S Fokas, P Theodoropoulos, Corinthia: K Farmaki, E Chaniotakis, P Theodoropoulou, K Karakonstantis, T Lygeros, Elis: D Pavlopoulou, I Lentzos, G Paraskeva, N Choundras, H Arvanitakis, G Stavropoulou, Mani: A Mariolis, P Lykousas, Messenia: I Adamopoulos, S Hideriotis, M Rentoulis, D Sotiropoulos, Laconia: M Tsironi, G Sakelariadis, A Vasimbosis, S Arahoviti, S Alevizou, P Andrianopoulos, Tsakonia: H Kontoleon, P Perdicari.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Stamatoyannopoulos, G., Bose, A., Teodosiadis, A. et al. Genetics of the peloponnesean populations and the theory of extinction of the medieval peloponnesean Greeks. Eur J Hum Genet 25, 637–645 (2017). https://doi.org/10.1038/ejhg.2017.18
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2017.18
This article is cited by
-
Identifying novel regulatory effects for clinically relevant genes through the study of the Greek population
BMC Genomics (2023)
-
Clinical pharmacogenomics in action: design, assessment and implementation of a novel pharmacogenetic panel supporting drug selection for diseases of the central nervous system (CNS)
Journal of Translational Medicine (2021)