Abstract
Meier-Gorlin syndrome (MGORS) is a rare disorder characterized by primordial dwarfism, microtia, and patellar aplasia/hypoplasia. Recessive mutations in ORC1, ORC4, ORC6, CDT1, CDC6, and CDC45, encoding members of the pre-replication (pre-RC) and pre-initiation (pre-IC) complexes, and heterozygous mutations in GMNN, a regulator of cell-cycle progression and DNA replication, have already been associated with this condition. We performed whole-exome sequencing (WES) in a patient with a clinical diagnosis of MGORS and identified biallelic variants in MCM5. This gene encodes a subunit of the replicative helicase complex, which represents a component of the pre-RC. Both variants, a missense substitution within a conserved domain critical for the helicase activity, and a single base deletion causing a frameshift and a premature stop codon, were predicted to be detrimental for the MCM5 function. Although variants of MCM5 have never been reported in specific human diseases, defect of this gene in zebrafish causes a phenotype of growth restriction overlapping the one associated with orc1 depletion. Complementation experiments in yeast showed that the plasmid carrying the missense variant was unable to rescue the lethal phenotype caused by mcm5 deletion. Moreover cell-cycle progression was delayed in patient’s cells, as already shown for mutations in the ORC1 gene. Altogether our findings support the role of MCM5 as a novel gene involved in MGORS, further emphasizing that this condition is caused by impaired DNA replication.
Similar content being viewed by others
Introduction
Meier-Gorlin syndrome (MGORS, MIM 224690) is a rare disorder characterized by severe intrauterine and post-natal growth retardation, bilateral microtia, and aplasia or hypoplasia of the patellae. Microcephaly is reported in 43% of patients.1 Further dysmorphic features include microstomia, full lips, highly arched or cleft palate, and micrognathia. Recessive variants in components of the pre-replication (pre-RC) or pre-initiation (pre-IC) complexes (ORC1, ORC4, ORC6, CDT1, CDC6, and CDC45), as well as de novo heterozygous variants in the DNA replication inhibitor protein GMNN, are associated to MGORS.2, 3, 4, 5, 6
We report a patient with a clinical diagnosis of MGORS in whom we detected, by whole-exome sequencing, biallelic variants in MCM5 (minichromosome maintenance complex component 5 [MIM 602696]), a gene encoding a member of the replicative helicase complex MCM2-7. Our findings further emphasize that MGORS is associated with impaired DNA replication.
Materials and methods
Clinical report
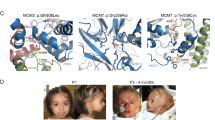
The patient was born at 41 weeks of gestation from non-consanguineous parents of Italian origin. Intrauterine growth restriction was documented after 36 weeks of gestation. At age 7 months, weight (6 kg, −2.9 SD) and length (61 cm, −3.3 SD) were both below the third centile and head circumference was 44.5 cm (0.5 SD). He presented with microstomia, thick lips, micrognathia, bilateral microtia, low set ears (Figure 1), and bilateral cryptorchidism. No feeding difficulties were reported. Abdominal ultrasound, following a persistent intestinal infection, revealed hypoplasia and ptosis of the left kidney. Psychomotor development and brain NMR were normal. Bilateral absence of the ossification centers of the patellae was noticed at age 20 months, when a clinical diagnosis of MGORS was established, and confirmed at a second evaluation at the age of 2 years and 6 months. Growth hormone levels were normal. Immunological investigations revealed a normal population of T- and NK-cells. At the age of 4 years and 8 months, patient’s weight and height were 12.5 kg (−2.7 SD) and 96.3 cm (−2.5 SD); head circumference was 51 cm (0.3 SD).
The parents’ height was 176 and 173 cm for the father and the mother, respectively. Patient’s growth curves and details, compared to MGORS individuals, are available in Supplementary Table S1 and Supplementary Figure S1.
Cytogenetic and whole-exome sequencing investigations
Written informed consent was obtained from the family according to the institutional review boards of IRCCS Fondazione Policlinico San Matteo, Pavia, Italy.
Karyotype, chromosome instability, and array-CGH analyses were performed as previously reported.7, 8 Spontaneous sister chromatid exchange analysis was done on patient’s and control’s lymphocytes.9 WES on the DNA of the patient and his parents (average coverage 75 ×) was performed by using a commercial target enrichment kit (SureSelectXT Clinical Research Exome, Agilent Technologies, Santa Clara, CA)on HiSeq2500 (paired-end 2 × 100 bp; Illumina, San Diego, CA, USA). After reads alignment, variant calling and annotation, and filtering for possible artifacts (see Supplementary Materials), candidate variants were compared with those reported in the following databases: 1000 Genomes Project, the Exome Sequencing Project, the Exome Aggregation Consortium (ExAC), and our internal database (189 exomes from patients with disorders not including growth impairment and their relatives). Non-synonymous variants or variants affecting splice-site acceptor/donor sites that were rare (MAF <0.01) or novel according to the above-mentioned databases were taken into consideration and analyzed in the trio according to both a de novo and a recessive hypothesis (both homozygous and compound heterozygous variants were considered for the latter). The resulting SNVs/InDels were visually inspected by Integrative Genomics Viewer IGV 1.210 to confirm the quality of the alignment. Relevant data were submitted to the Leiden Open Variation Database at www.lovd.nl/MCM5 (patient ID #00081271).
Further details on ‘Materials and Methods’ section are provided in Supplementary Materials.
Results and discussion
The patient’s karyotype was 46,XY, and array-CGH analysis resulted negative. WES identified about 11 000 non-synonymous or splice-sites variants in the proband, 434 being rare or novel (Supplementary Table S2). After filtering in the trio we remained with eight possible compound heterozygous variants affecting four genes (Supplementary Table S3). As expected from the family history, no candidate homozygous variants were detected. Moreover, no candidate de novo variants were identified. None of the filtered variants was in already known MGORS genes.
MCM5 appeared to be a strong candidate, because the encoded protein is part of the replicative helicase complex MCM2-7. MCM2-7 is recruited on the replication origins in the early G1 phase by the ORC1-6 complex through CDT1 and CDC6 (origin licensing). This pre-RC is further activated at the G1-S-phase transition by cell-cycle dependent kinases (CDK and DKK), allowing the assembly of the pre-IC complex, including CDC45, the unwinding of the DNA, and finally, its replication.6, 11 Genes encoding for multiple components of this process have already been found mutated in MGORS patients.2, 3, 5, 6, 12 Moreover, although MCM5 alterations have never been associated with human diseases, spontaneous mcm5 mutants13 and morphant models2 in zebrafish strictly overlap the phenotype of growth restriction caused by orc1 depletion.
The two variants affecting MCM5 in our patient (Figure 2), namely c.850_851delAG, p.(Arg284Glyfs*49), of paternal origin, and c.1397C>T (p.Thr466Ile), inherited from the mother, were both predicted to be damaging with high confidence by the in silico tools we used (Supplementary Table S4).
MCM5 gene variants identified in our case. (a) The two panels show the IGV screenshots relative to the c.850_851del (upper panel) and the c.1397C>T (lower panel) variants in patient’s DNA. Gray bars represent the mapped reads aligned to the reference genome, which sequence is shown below (colored). A coverage plot is displayed in the upper part of each panel. (b) The results of Sanger sequencing of exons 7 (upper panel) and 11 (lower panel) of the MCM5 gene are shown for the proband (Pb), his father (F) and mother (M). The c.850_851del deletion (upper panel) was detected in the paternal DNA, whereas the c.1397C>T variant (lower panel) was maternally inherited. The altered nucleotides are indicated by arrows. MCM5 RefSeq accession number: NM_006739.3.
The c.850_851delAG change was predicted to result in non-sense mediated mRNA decay (NMD). We assessed the relative abundance of the two transcripts by deep-sequencing of a 519 bp fragment encompassing the c.1397C>T variant in the cDNA from lymphoblastoid cell lines (LCLs) of the patient and his parents. In the patient’s sample the c.1397C>T variant (corresponding to r.1397c>u) was called by 77% of the reads, whereas the allele harboring the frameshift variant was represented by 23% only of the reads, with a shift from a 1:1 to a 3:1 ratio in favor of the transcript bearing the c.1397C>T variant (Supplementary Table S5). This indicated a selective partial degradation of the transcript bearing the c.850_851delAG variant, supporting the hypothesis of NMD. Western blot (Supplementary Figure S2) analysis also showed a reduction of the MCM5 protein in LCLs lysates from the patient and his father, carrying the frameshift variant. MCM5 was only barely detectable in the nuclear (insoluble) fraction of fibroblast cells lysates from the patient compared to a control. This finding might be explained by an impairment of the MCM2-75p.T466I to stably maintain its association with the chromatin, as suggested for mutations affecting the same domain in S. cerevisiae.11 Consistent with this hypothesis, we observed a comparable decrease in the levels of chromatin-bound MCM2 (Supplementary Figure S3).
The c.1397C>T variant (p.Thr466Ile), affects a region highly conserved among orthologous MCM5 proteins and different members of the MCM2-7 complex (Supplementary Figure S4). This region is also conserved in the SsoMCM protein from archaea Sulfolobus solfataricus,14 which X-ray structure was used to do in silico structural analysis of p.Thr466Ile in human MCM5. The p.Thr466Ile substitution affects a conserved residue located in the pre-sensor 1 domain (PS1) of MCM5 (Supplementary Figure S5), one of the three β-hairpin structures of the C-terminal domain that are critical for the activity of MCM proteins.14, 15 This domain is also highly conserved in all the MCM subunits of S. cerevisiae, where mutations in the PS1 impair the helicase activity of the MCM2-7 complex.11, 16
The pathogenicity of the p.Thr466Ile variant (corresponding to p.Thr501Ile in yeast) was further supported by a yeast-based functional complementation assay17 in S. cerevisiae (strain W303) carrying the mcm5 deletion, as reported in Supplementary Materials. Transformation with the plasmid containing the wild-type mcm5 effectively rescued the growth phenotype of the tetrad, whereas the plasmid carrying the p.Thr501Ile mutant had no effect, indicating that the mutation is detrimental for Mcm5p protein function (Figure 3).
Functional complementation in yeast. (a) Alignment of human and yeast MCM5 protein sequences: the mutated threonine residue is underlined. (b) A copy of the mcm5 gene was inactivated in a diploid W303 yeast by homologous recombination with a KANMX4 cassette (which confers resistance to geneticin). The strains carrying the heterozygous deletion were left to sporulate on solid medium for 4 days at 25 °C; 10 tetrads were dissected and plated in rich medium at 30° for 3 days. Four representative tetrads (1–4) are shown: only two spores for each tetrad survived and none of them could grow in medium containing geneticin (YPD +G418, on the right), confirming that these haploid cells carried the wild-type allele. (c) Diploid yeasts, heterozygous for mcm5 inactivation, were transformed by plasmids containing either the wild type (pCM189-ΔCYC1-mcm5-WT) or the mutated (pCM189-ΔCYC1-mcm5-T501I) mcm5, and carrying a URA3 selection cassette. Transformants were selected in synthetic medium lacking uracil (SM–URA) and then left to sporulate; 16 tetrads were dissected and four representative tetrads for each plasmid are shown (1–4 wild type; 5–8 mutant). Each colony was then streaked in medium lacking uracil (SM–URA) to confirm the presence of the plasmid and then in geneticin-containing medium (YPD +G418) to confirm the mcm5 genotype. Transformation by the wild-type mcm5 restored a normal segregation pattern: all the four spores of each tetrad (1–4) could grow in SM–URA, both the two carrying the inactivated allele and the two carrying the wild-type mcm5. The latter could no more survive in geneticin-containing medium (YPD +G418; spores: 1B, 1D, 2A, 2B, 3A, 3C, 4B, 4C). In contrast, only 50% of those transformed with mutant mcm5 survived. All the surviving spores (5A, 5B, 6A, 6B, 7B, 7D, 8B, and 8C) carried the wild-type allele, as demonstrated by their geneticine-sensitivity (YPD+G418). This indicated that the Thr501Ile abolishes the function of mcm5.
To date, the only other member of the MCM2-7 complex found mutated in humans is MCM4 (MIM 602638). A single ancestral variant of this gene (NM_005914.3:c.71-2A>G), resulting in a N-terminal truncated protein, was associated at homozygous state with short stature, adrenal insufficiency, natural killer cell deficiency, and genomic instability, in multiple families from an isolated Irish population.18, 19 The reported phenotype, with the exception of short stature, is rather different in respect to MGORS. Moreover our patient did not present any sign of immunological defects, even after extensive investigations, and this condition has never been reported in MGORS. These differences may be explained by the specific effect of the c.71-2A>G on the MCM4 protein, or by the specific genetic background in which this variant was found.
We have also tested patient’s cells for chromosome instability and for signs of centrosomes abnormalities, as reported for other primordial dwarfisms20, 21 and, possibly, for some ORC1 mutations.22 However we did not identify in our patient either signs of chromosome instability, or increased number of cells with amplified centrosomes or micronuclei (Supplementary Table S6 and Supplementary Figure S6).
A slow S-phase progression has been reported in LCLs from ORC1 mutated MGORS2. We thus investigated by BrdU pulse-chase analysis both LCLs and primary skin fibroblasts from our patient. A significant delay was observed in both cell types (Supplementary Figures S7 and S8).
We also tested cells hypersensitivity to replicative stress, a feature reported in association to a reduction in the helicase complex.23 To this purpose, we treated patient’s and control’s LCLs with the replication inhibitor hydroxyurea. A reduced percentage of patient’s cells proceeded towards the S-phase (Supplementary Figure S9). This is possibly explained by the impairment of the so-called ‘dormant origins’ to be activated, as previously suggested by Ge et al.23
In summary, we report MCM5 as a novel gene associated with MGORS. Although different pathogenetic mechanism for MGORS were proposed, the more likely appears to be an impairment of DNA replication initiation, since all the genes thus far associated with this disease are involved in this pathway.
References
de Munnik SA, Hoefsloot EH, Roukema J et al: Meier-Gorlin syndrome. Orphanet J Rare Dis 2015; 10: 114.
Bicknell LS, Walker S, Klingseisen A et al: Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat Genet 2011; 43: 350–355.
Guernsey DL, Matsuoka M, Jiang H et al: Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat Genet 2011; 43: 360–364.
de Munnik SA, Bicknell LS, Aftimos S et al: Meier-Gorlin syndrome genotype-phenotype studies: 35 individuals with pre-replication complex gene mutations and 10 without molecular diagnosis. Eur J Hum Genet 2012; 20: 598–606.
Burrage LC, Charng WL, Eldomery MK et al: De Novo GMNN mutations cause autosomal-dominant primordial dwarfism associated with Meier-Gorlin syndrome. Am J Hum Genet 2015; 97: 904–913.
Fenwick AL, Kliszczak M, Cooper F et al: Mutations in CDC45, encoding an essential component of the pre-initiation complex, cause meier-gorlin syndrome and craniosynostosis. Am J Hum Genet 2016; 99: 125–138.
Vetro A, Iascone M, Limongelli I et al: Loss-of-function FANCL mutations associate with severe fanconi anemia overlapping the VACTERL association. Hum Mutat 2015; 36: 562–568.
Vetro A, Manolakos E, Petersen MB et al: Unexpected results in the constitution of small supernumerary marker chromosomes. Eur J Med Genet 2012; 55: 185–190.
German J, Schonberg S, Louie E, Chaganti RS : Bloom's syndrome. IV. Sister-chromatid exchanges in lymphocytes. Am J Hum Genet 1977; 29: 248–255.
Thorvaldsdottir H, Robinson JT, Mesirov JP : Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013; 14: 178–192.
Ramey CJ, Sclafani RA : Functional conservation of the pre-sensor one beta-finger hairpin (PS1-hp) structures in mini-chromosome maintenance proteins of Saccharomyces cerevisiae and archaea. G3 (Bethesda) 2014; 4: 1319–1326.
Bicknell LS, Bongers EMHF, Leitch A et al: Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat Genet 2011; 43: 356–359.
Ryu S, Holzschuh J, Erhardt S, Ettl AK, Driever W : Depletion of minichromosome maintenance protein 5 in the zebrafish retina causes cell-cycle defect and apoptosis. Proc Natl Acad Sci USA 2005; 102: 18467–18472.
Brewster AS, Wang G, Yu X et al: Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc Natl Acad Sci USA 2008; 105: 20191–20196.
McGeoch AT, Trakselis MA, Laskey RA, Bell SD : Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat Struct Mol Biol 2005; 12: 756–762.
Lam SK, Ma X, Sing TL, Shilton BH, Brandl CJ, Davey MJ : The PS1 hairpin of Mcm3 is essential for viability and for DNA unwinding in vitro. PLoS One 2013; 8: e82177.
Trevisson E, Burlina A, Doimo M et al: Functional complementation in yeast allows molecular characterization of missense argininosuccinate lyase mutations. J Biol Chem 2009; 284: 28926–28934.
Gineau L, Cognet C, Kara N et al: Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. J Clin Invest 2012; 122: 821–832.
Hughes CR, Guasti L, Meimaridou E et al: MCM4 mutation causes adrenal failure, short stature, and natural killer cell deficiency in humans. J Clin Invest 2012; 122: 814–820.
Mirzaa GM, Vitre B, Carpenter G et al: Mutations in CENPE define a novel kinetochore-centromeric mechanism for microcephalic primordial dwarfism. Hum Genet 2014; 133: 1023–1039.
Rauch A, Thiel CT, Schindler D et al: Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science 2008; 319: 816–819.
Hossain M, Stillman B : Meier-Gorlin syndrome mutations disrupt an Orc1 CDK inhibitory domain and cause centrosome reduplication. Genes Dev 2012; 26: 1797–1810.
Ge XQ, Jackson DA, Blow JJ : Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev 2007; 21: 3331–3341.
Acknowledgements
AV benefits of a research position granted by the University of Pavia in the context of the strategic plan: Towards a governance model for international migration: an interdisciplinary and diachronic perspective (MIGRAT-IN-G). LS is supported by Telethon Italy Grant GGP13222, a grant from Fondazione CARIPARO, and University of Padova Grant CPDA123573/12; OZ is supported by Telethon Italy Grant GGP13060. The URLs for data presented herein are as follows: Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA, release ESP6500SI-V2 (http://evs.gs.washington.edu/EVS); ExAC (http://exac.broadinstitute.org); Integrative Genomics Viewer (IGV),http://www.broadinstitute.org/igv/Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/ Phyre2 webserver http://www.sbg.bio.ic.ac.uk/phyre2/ Protein Data Bank (PDB), http://www.rcsb.org/pdb/home/home.do; and RefSeq, http://www.ncbi.nlm.nih.gov/RefSeq.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Vetro, A., Savasta, S., Russo Raucci, A. et al. MCM5: a new actor in the link between DNA replication and Meier-Gorlin syndrome. Eur J Hum Genet 25, 646–650 (2017). https://doi.org/10.1038/ejhg.2017.5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2017.5
This article is cited by
-
The expanding genetic and clinical landscape associated with Meier-Gorlin syndrome
European Journal of Human Genetics (2023)
-
Comment on: “The expanding genetic and clinical landscape associated with Meier-Gorlin syndrome” by Nielsen-Dandoroff et al.
European Journal of Human Genetics (2023)
-
Unwinding the Role of the CMG Helicase in Inborn Errors of Immunity
Journal of Clinical Immunology (2023)
-
Enhanced MCM5 Level Predicts Bad Prognosis in Acute Myeloid Leukemia
Molecular Biotechnology (2023)
-
RNA Sequencing and Bioinformatics Analysis Reveals the Downregulation of DNA Replication Genes by Morindone in Colorectal Cancer Cells
Applied Biochemistry and Biotechnology (2023)