Abstract
Anophthalmia and microphthalmia (A/M) are developmental ocular malformations defined as the complete absence or reduction in size of the eye. A/M is a highly heterogeneous disorder with SOX2 and FOXE3 playing major roles in dominant and recessive pedigrees, respectively; however, the majority of cases lack a genetic etiology. We analyzed 28 probands affected with A/M spectrum (without mutations in SOX2/FOXE3) by whole-exome sequencing. Analysis of 83 known A/M factors identified pathogenic/likely pathogenic variants in PAX6, OTX2 and NDP in three patients. A novel heterozygous likely pathogenic variant in PAX6, c.767T>C, p.(Val256Ala), was identified in two brothers with bilateral microphthalmia, coloboma, primary aphakia, iris hypoplasia, sclerocornea and congenital glaucoma; the unaffected mother appears to be a mosaic carrier. While A/M has been reported as a rare feature, this is the first report of congenital primary aphakia in association with PAX6 and the identified allele represents the first variant in the PAX6 homeodomain to be associated with A/M. A novel pathogenic variant in OTX2, c.651delC, p.(Thr218Hisfs*76), in a patient with syndromic bilateral anophthalmia and a hemizygous pathogenic variant in NDP, c.293 C>T, p.(Pro98Leu), in two brothers with isolated bilateral microphthalmia and sclerocornea were also identified. Pathogenic/likely pathogenic variants were not discovered in the 25 remaining A/M cases. This study underscores the utility of whole-exome sequencing for identification of causative mutations in highly variable ocular phenotypes as well as the extreme genetic heterogeneity of A/M conditions.
Similar content being viewed by others
Introduction
Anophthalmia and microphthalmia (A/M) are developmental ocular malformations in which the eye fails to form or is smaller than normal. Microphthalmia is often associated with additional ocular anomalies, most commonly coloboma or cataract;1, 2 approximately one-third of microphthalmia includes coloboma (an optic fissure closure defect, OFCD) and a little more than a third of non-OFCD microphthalmia includes cataract.2 A/M conditions have been estimated to affect 2–6 in 50 000 live births.1, 2, 3 A/M can be an isolated ocular condition but approximately one-third of all A/M cases are syndromic, associated with features such as craniofacial, genital, skeletal, brain, renal and cardiac abnormalities;2, 4, 5 neurological anomalies appear to be most common in patients with anophthalmia while urological and genital anomalies were most prevalent in patients with OFCD conditions (with or without microphthalmia).2 A specific pattern of associated features can provide a clinical diagnosis in approximately 25% of cases.6
A/M is a genetically heterogeneous disorder with dominant, recessive, and X-linked inheritance patterns reported;6 pathogenic variants in SOX2 and OTX2 represent the most common causes of dominant A/M while FOXE3 is the most common source of recessive microphthalmia, each explaining 8–15% of cases.7, 8, 9, 10 Additional mutations have been found in PAX6, STRA6, ALDH1A3, RARB, VSX2, RAX and BMP4, along with numerous other genes; however, the majority of A/M cases do not have a genetic explanation at this time.1 Screening for intragenic or copy number mutations in seven (or eight) known genes in patients with A/M successfully identified a causative mutation in 21–30% of cases.10, 11 Whereas some genes are specifically associated with either OFCD (eg, STRA6) or non-OFCD (eg, FOXE3) conditions, many genes can cause either type of ocular disorder (eg, SOX2, OTX2, VSX2); in addition, many genes can be associated with both syndromic and isolated A/M.2 The majority of cases are sporadic and the occurrence of de novo mutations, mosaicism and incomplete penetrance makes prediction of the inheritance pattern difficult.4, 5 Empiric recurrence risk to siblings is 6–13%5 but the actual risk varies between 0–50% depending on the cause; thus, establishing a genetic diagnosis is of great value to families. Beyond defining recurrence risk information, identification of the genetic etiology may guide clinical management for the affected child.12, 13, 14 Owing to the number of genes involved, the variability of associated phenotypes, and the large proportion of unexplained cases, whole-exome sequencing (WES) has been shown to be effective in both screening known genes and identifying new causative factors in families with A/M.14, 15, 16, 17
Materials and methods
Ethics statement
The human study was approved by the Institutional Review Board of the Children’s Hospital of Wisconsin with written informed consent obtained from each participant and/or their legal representative, as appropriate.
Human patients
DNA samples from 28 probands affected with microphthalmia, anophthalmia, or coloboma were submitted for WES; all of the DNA samples were extracted from blood using standard protocols with the exception of 1 sample extracted from amniocytes and 1 sample extracted from fibroblasts. Of these, 5 had anophthalmia (unilateral in 1 with ASD of the contralateral eye), 22 had microphthalmia and 1 had isolated coloboma; 22 probands had bilateral ocular anomalies. Overall, 15 cases were syndromic (mild in 4) and the remaining 13 cases had isolated ocular anomalies. The most common ocular feature in addition to A/M was coloboma (12 cases). Familial ocular anomalies were noted in 9 cases and the remaining 19 were sporadic. Of the 28 families, 15 were of Caucasian (European), 5 of Caucasian (Middle Eastern), 2 of Asian, 4 of Hispanic, 1 of African American and 1 of Native American/Caucasian race/ethnicity. All 28 samples were negative for disease-causing variants in SOX2 and FOXE3 by Sanger sequencing and most were also screened for variants in OTX2, BMP4, and VSX2 as well as array-CGH for genomic deletions/duplications; 8 previously published cases with disease-causing variants in these genes were excluded from the WES cohort.7, 8, 9, 18, 19, 20
Whole-exome sequencing and variant confirmation
WES was performed by Perkin Elmer, Inc (Branford, CT, USA) or Axeq (Rockville, MD, USA); exome capture was performed with the Agilent Sure Select v4 + UTR (Santa Clara, CA, USA) and 100 base pair paired end sequencing was performed using the Illumina HiSeq 2000 (San Diego, CA, USA). The data alignments were performed by the sequencing center using the Burrows-Wheeler Aligner (BWA) followed by variant calling with either the Genome Analysis Toolkit (GATK v2.20, Broad Institute, Cambridge, MA, USA) pipeline available through Perkin Elmer or the Sequence Alignment/Map (SAMtools) pipeline through Axeq. To examine genes previously associated with A/M or coloboma, a candidate list of 83 genes was utilized, including COL4A1 and MAB21L2 recently identified to have a role in A/M through exome sequencing by our and other groups14, 15, 16 (Supplementary Table 1). Data were analyzed using the SNP & Variation Suite (SVS; Golden Helix, Bozeman, MT, USA). The frequency in general populations was determined based on the Database of Single Nucleotide Variants (dbSNP, http://www.ncbi.nlm.nih.gov/snp/), 1000 genomes sequencing project (http://www.1000genomes.org/), the National Heart Lung and Blood Institute Exome Variant Server database (EVS, http://evs.gs.washington.edu/EVS/) and the ExAC browser (http://exac.broadinstitute.org/). The predicted functional impact of missense variants was ascertained through SIFT, PolyPhen, MutationTaster, MutationAssessor, FATHMM and Radial SVM prediction algorithms as well as GERP++ and PhyloP conservation scores accessed through SVS utilizing data from dbNSFP.21 Once variants of interest were identified, primers flanking the variant sites were designed and genomic DNA was amplified in probands and all available family members to determine inheritance (Supplementary Table 2). PCR products were sequenced bidirectionaly using Big Dye Terminator chemistry and ABI 3730XL sequencer (Applied Biosystems/Life Technologies, Carlsbad, CA, USA). Sequences were reviewed manually and using Mutation Surveyor (SoftGenetics, State College, PA, USA). Variants were classified as pathogenic, likely pathogenic, uncertain significance, likely benign or benign using the recently published consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology for interpretation of sequence variants which considers population data, computational/predictive data, and functional data, as well as other genetic data available for the proband/family.22 Confirmed variants were submitted to the LOVD 3.0 shared installation databases: (http://databases.lovd.nl/shared/genes/PAX6 (patient ID 37525), http://databases.lovd.nl/shared/genes/otx2 (patient ID 37524) and http://databases.lovd.nl/shared/genes/ndp (patient ID 37523)).
Results
Known A/M gene analysis in anophthalmia/microphthalmia probands
Mean coverage of all 28 probands was 67.5±19.0 with 95.2%±2.2% of bases with >10 × coverage. Analysis of 83 known A/M genes identified 2235 variants; of these 592 were located either inside or within 15 base pairs of an exon. Of these 592 variants, 388 had an allele frequency >1% in the general population and thus were considered benign. There were 68 rare nonsynonymous variants (each present in one or two probands); 56 were observed in the general population but at a frequency below 1% and 12 were novel. Among these variants, 2 were frameshift and 66 were missense. Functional significance analysis of missense variants identified 4 variants which were categorized as damaging by 6 out of 6 online prediction tools (SIFT, PolyPhen, MutationTaster, MutationAssessor, FATHMM and Radial SVM), 8 by 5 tools, 6 by 4, 12 by 3, 8 by 2 and 20 by only 1 tool; 8 variants were called benign by all 6 programs (Supplementary Table 3). Pathogenic/likely pathogenic heterozygous variants in the PAX6, OTX2 and NDP genes were identified in three probands and determined to be the cause of the A/M in these probands (Table 1).
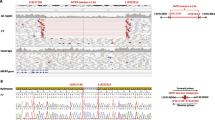
A novel heterozygous likely pathogenic variant in PAX6, c.767T>C, p.(Val256Ala), was identified in Patient 1, a 6-year-old Caucasian (European) male affected with bilateral microphthalmia, papillary coloboma, primary aphakia, iris hypoplasia, sclerocornea and congenital glaucoma; extraocular anomalies include low-set prominent ears and microcephaly (Table 1; Figure 1). The variant affects a highly conserved valine residue (GERP++ score 5.53 and PhyloP score 2.216) in the homeodomain region of PAX6 and is predicted to be damaging by all six programs; the variant is not reported in dbSNP, 1000 Genomes, EVS or ExAC databases. The variant has not been previously reported based on searches of the scientific literature and the Leiden Open Variation Database (LOVD) (http://lsdb.hgu.mrc.ac.uk/home.php?select_db=PAX6). The patient’s 4-year-old brother is similarly affected with bilateral microphthalmia, iris hypoplasia, sclerocornea and glaucoma, unilateral optic nerve coloboma, unilateral aphakia with subluxation of the lens in the other eye, asymmetric facies and a history of mild developmental delays which resolved; he was found to carry the same variant. Both of the parents are unaffected and analysis of their DNA (extracted from blood) by Sanger sequencing identified the mother as a likely mosaic carrier of the c.767T>C, p.(Val256Ala) pathogenic variant; no other tissue was available from the mother for comparison (Figure 1).
PAX6 pathogenic variant. (a) Pedigree of Patient 1. Patient 1 (proband) is indicated with an arrow; genotype of each individual is listed below the corresponding symbol. (b) DNA chromatograms for Patient 1 and family members are shown with the position of the c.767T>C, p.(Val256Ala) mutation indicated with red arrows. Please note a smaller ‘C’ peak at position c.767 in addition to the wild-type ‘T’ in DNA sample of the unaffected mother, suggesting that she is likely to be mosaic for this mutation. (c) Amino-acid alignment of the PAX6 homeodomain from different species. The alignment utilized KALIGN (http://www.ebi.ac.uk/Tools/msa/kalign) and the following reference sequences: AAK95849.1 (human PAX6), AFE78491.1 (Rhesus Pax6), AAI16039.1 (cow Pax6), AAH36957.1 (mouse Pax6), AAI28742.1 (rat Pax6), BAA23004.1 (chicken Pax6), NP_571379.1 and AAC96095.1 (zebrafish pax6a and pax6b, correspondingly; only one HD sequence is shown because it is identical between the two homologs), NM_079889.3 (eyeless (ey); Drosophila melanogaster), AAF59395.4 (twin of eyeless (toy), D. melanogaster), AAA82991.1 (mab-18, Caenorhabditis elegans). The position 256 of the homeodomain is shown in bold and is occupied by a valine residue in all available reference sequences and alanine in Patient 1 and his affected brother. The positions of the previously reported PAX6 mutations affecting the HD are underlined in the human reference sequence. (d) Schematic representation of the PAX6 protein. The positions of PAX6 domains are indicated with different colors and numbers at the bottom of the drawing; PD (paired domain), LNK (linker region), HD (homeodomain), PST (proline–serine–threonine-rich transactivation domain); the position of the protein region encoded by the alternatively spliced exon 5a is also specified. The position of the mutation in Patient 1 is indicated with a red arrow; the positions of published mutations involving the homeodomain are shown with grey arrows; the positions of previously reported A/M mutations are marked with black arrows (for missense) or black arrows with an asterisk (nonsense); dominant (heterozygous) mutations are shown at the top and compound heterozygous mutations (with allelic combinations being connected with a line) on the bottom of the drawing. A full color version of this figure is available at the European Journal of Human Genetics journal online.
A novel heterozygous pathogenic variant in OTX2, c.651delC, p.(Thr218Hisfs*76), was identified in Patient 2, a 6-day-old Hispanic (El Salvador) male with bilateral clinical anophthalmia (Table 1; Figure 2). Head CT (computed tomography) scanning noted absent globes but presence of optic nerves; an MRI of the brain and orbits showed left anophthalmia and right microphthalmia vs anophthalmia and no anomalies of the brain. Birth weight was normal (3.7 kg) and a renal ultrasound performed in the newborn period was also normal. Follow-up that occurred after identification of the OTX2 variant revealed new extraocular features. His height was 85 cm (<3rd centile), his weight was 14.5 kg, (25–50th centile), and his head circumference was 48 cm (10–25th centile) at the age of 3 years and 3 months. Dysmorphic features included eversion of the left upper lid, blunt grooved uvula, long chin, depressed nasal tip and large and fleshy ears along with tapered fingers, joint hypermobility throughout, one hypopigmented macule on the belly, impalpable left testis and small scrotum. Intellectual disability was evident and an autistic spectrum disorder was suspected; he was not able to cruise or walk, used only single words, did not follow directions, did not initiate interaction and demonstrated repetitive movements. At the age of 5, he was diagnosed with cerebral palsy, hyperactive deep tendon reflexes, and abnormal cerebellar function. Patient 2’s parents and three siblings are unaffected but were not available for genetic testing. The variant is located in the last exon of OTX2 and thus is not expected to undergo nonsense mediated decay; this variant is absent in general populations (dbSNP, 1000 Genomes, EVS, EXAC).
OTX2 pathogenic variant. (a) Pedigree of Patient 2. Patient 2 is indicated with an arrow and his OTX2 genotype is listed. (b) DNA chromatograms for Patient 2 with the position of the c.651delC, p.(Thr218Hisfs*76) mutation indicated with a red arrow. (c) Schematic representation of the OTX2 protein. The positions of the OTX2 domains are indicated with different colors and numbers at the bottom of the drawing; HD (homeodomain), SGQFTP and SIWSPA sequences that are highly conserved within the OTX family and two OTX2 tail motifs that are required for the transactivation activity. The position of the mutation in Patient 2 is indicated with a red arrow. A full color version of this figure is available at the European Journal of Human Genetics journal online.
A hemizygous pathogenic variant in NDP, c.293 C>T, p.(Pro98Leu), was discovered in Patient 3, an 18-year-old Caucasian (Iran) male affected with isolated bilateral microphthalmia and sclerocornea (Figure 3). His similarly affected 14-year-old brother is also hemizygous for this variant and his unaffected mother is a heterozygous carrier. Neither of the brothers have hearing loss, cognitive impairment or psychiatric/behavioral concerns.
NDP pathogenic variant. (a) Pedigree of Patient 3. Patient 3 is indicated with an arrow and NDP genotypes are listed for all examined family members. (b) DNA chromatograms for Patient 3 and his family members with the position of the c.293 C>T, p.(Pro98Leu) mutation indicated with a red arrow. (c) Amino-acid alignment of the NDP region around the p.(Pro98Leu) mutation from various species. The following reference sequences were used: human (AK313409.1), cow (BC112738.1), mouse (BC090623.1), chicken (NM_001278087.1), Xenopus (NM_001161397.1) and zebrafish (XM_009304808.1). The position 98 is shown in bold and is occupied by a proline residue in all available reference sequences and leucine in Patient 3 and his affected brother. (d) Schematic representation of the NDP protein. The positions of the NDP domains are indicated with different colors and numbers at the bottom of the drawing; SP (signal peptide), CTCK (C-terminal cystine knot-like domain). The position of the mutation in Patient 3 is indicated with a red arrow. A full color version of this figure is available at the European Journal of Human Genetics journal online.
Eight novel or rare (present in <15 controls) variants in dominant genes were identified and considered to be variants of uncertain significance (GLI2 (Patient 4), GLI3 (Patient 5), CHD7 (Patient 8 and 18), PTCH1 (Patient 19), SALL1(Patient 22), FOXL2 (Patient 19), and BMP7 (Patient 25)) since there was not enough evidence to classify them as either likely benign or likely pathogenic; in addition, the patient phenotypes were not consistent with the previously reported syndromes for well-characterized genes (Supplementary Table 3). The remaining variants were considered unlikely to be causative in the proband based on their classification as benign/likely benign using the ACMG recommendations or lack of a second variant (for a gene with a known autosomal recessive inheritance pattern).
Discussion
In this manuscript we report exome sequencing analysis of 83 factors known to be associated with anophthalmia/microphthalmia and/or coloboma in 28 patients affected with these conditions and prescreened for variants in major causative genes (using gene-specific Sanger sequencing and CNV assays). Pathogenic/likely pathogenic variants in known A/M factors were identified in three affected individuals in this prescreened group (11%); this is lower than the 20–30% detection rate previously reported by other groups10, 11 which is likely due to the prior exclusion of major contributors, most notably SOX2 and FOXE3, in our cohort. The lack of causative variants in known ocular factors in the majority of the prescreened cohort (25 patients; 89%) further emphasizes the extreme genetic heterogeneity of A/M phenotypes. Though it is possible that some nucleotide variants located within intronic or regulatory regions or large deletions/duplications of known factors were missed by WES,23 contribution from additional, yet to be discovered, genes to A/M conditions seems likely. The identification of variants of uncertain significance in seven patients highlights the challenge of interpretation of whole-exome data, even within a restricted list of known disease genes. The identified pathogenic/likely pathogenic variants broaden the phenotypic spectra associated with the corresponding genes; additionally, two of the identified disease-causing variants are novel.
PAX6 encodes a transcription factor consisting of a bipartite paired domain and a homeodomain, both responsible for DNA binding, and a proline/serine/threonine-rich region involved in transactivation. Pax6 is initially expressed in the anterior neural plate and by E8.0 is detected in the lens field of the head surface ectoderm and optic pit, which gives rise to the optic vesicle; expression is maintained throughout life in the cornea, lens epithelium and the retina.24 PAX6 was first identified as a candidate gene responsible for WAGR syndrome through deletion analysis and subsequent studies identified truncation, missense and splicing disease-causing variants in isolated aniridia cases.25 Missense variants represent the minority of pathogenic alleles, but result in the most variable phenotypes (50 missense variants are listed in the PAX6 LOVD; two-thirds of them are located within the paired domain).25 About half of reported missense variants cause aniridia, whereas the second half result in iris hypoplasia, cataracts, foveal hypoplasia, Peters anomaly and other phenotypes. Anophthalmia/microphthalmia is part of the PAX6-associated phenotypic spectrum but reported in a very small number of cases: five heterozygous missense p.(Arg19Pro), p.(Arg26Gly), p.(Leu46Pro), p.(V54D)/(p.(5aV7D), p.(Gln378Arg), one heterozygous nonsense variant p.(Arg203*), as well as a case with a heterozygous p.(Arg38Trp) PAX6 variant in combination with an OTX2 nonsense allele have been reported in patients with microphthalmia along with corneal opacity, cataract and/or iris hypoplasia/aniridia;26, 27, 28, 29, 30, 31, 32 four individuals with compound heterozygous PAX6 disease-causing variants, p.[(Arg38Trp)];[(Arg240*)], p.[(Arg103*)];[(Ser353*)], p.[(Arg38Glyfs*16)];[(Cys40_Ser43del)] (in two siblings), were reported with anophthalmia/severe microphthalmia and severe brain anomalies (lethal in all but the first patient).33, 34, 35 No additional deleterious alleles in PAX6 or other known ocular genes have been identified in our patient, although a contribution from new ocular factors, yet to be discovered, cannot be ruled out. Overall, the presented variant fits well with the previously reported cases but represents the first variant in the homeodomain to be associated with A/M and, to the best of our knowledge, the first report of congenital primary aphakia in association with a PAX6 variant.
The homeodomain of PAX6 has the smallest number of reported variants relative to other regions. The PAX6 LOVD lists only six missense variants in the homeodomain (p.(Arg214Gly), p.(Arg214Ser), p.(Ser216Pro), p.(Arg242Thr), p.(Gln255His), p.(Phe258Ser)) with phenotypes including mild aniridia, iris hypoplasia, iridocorneal adhesions, chorioretinal and optic nerve coloboma, persistent hyperplastic primary vitreous, cataracts, glaucoma, corneal clouding and sclerocornea but not microphthalmia.5, 30, 36, 37, 38, 39 Therefore, the phenotype described in Patient 1 and his affected brother represents the most severe manifestation of PAX6 homeodomain missense variants. Multiple Pax6 mutant alleles in mice have been reported resulting in small eye and other ocular phenotypes.25 Most relevant to this study is the Pax6 variant c. 971T>A, p.(Val270Glu) that was identified through an ENU mutagenesis project affecting the same valine residue that is mutated in Patient 1.40 The mice demonstrated normal size eye, corneal dimple and an irregular pupil.40
The OTX2 gene encodes a transcription factor critical for forebrain and eye development.41 Previous families have shown a dominant pattern for OTX2 loss-of-function variants, with frequent de novo changes.1, 9 The OTX2 protein contains a homeodomain, responsible for DNA binding, SGQFTP and SIWSPA motifs involved in protein–protein interactions, and two C-terminal tandem OTX-tail motifs responsible for transactivation. The c.651delC variant is predicted to escape NMD and represents the most C-terminal truncation allele reported to date; the encoded mutant protein p.(Thr218Hisfs*76) may exhibit dominant-negative effects as it is expected to retain the homeodomain and protein interaction domains, but to lack the transactivation domain. The severe ocular phenotype observed in Patient 2, as well as the additional systemic defects (short stature, developmental delay) are consistent with previously described OTX2-positive cases, particularly the ones retaining the homeodomain.9, 42, 43 Several less common/not previously reported features such as autism spectrum disorder, joint hypermobility and cerebral palsy were also present. Although autism spectrum disorder and joint laxity are not commonly recognized features of OTX2 microphthalmia syndrome, both were described in at least one other patient.9, 44 Cerebellar abnormalities have not been previously noted in patients with OTX2 disease-causing variants (but other brain anomalies such as agenesis of the corpus callosum and structural pituitary defects have been recurrently reported42). At the same time, the Otx factors were shown to be critical for cerebellar regionalization in animal models45, 46 and OTX2 expression has been detected in the cerebellum as well as other parts (archicortex, diencephalon, rostral brain stem) of the early developing brain in humans.47 In addition, OTX2 has been shown to act as an oncogene in medulloblastoma, the most common pediatric malignant brain tumor which is always located in the cerebellum.48, 49
The NDP gene encodes norrin, a protein that contains a cysteine knot motif similar to members of the TGF-β family and acts in the canonical Wnt signaling pathway. NDP variants have been identified as the cause of Norrie Disease, an X-linked recessive disorder characterized by congenital blindness due to fibrous or vascular changes in the retina with leukocoria along with hearing loss in most patients and cognitive/behavioral abnormalities in 30–50%,50, 51 as well as X-linked familial exudative vitreoretinopathy, associated with abnormal vascularization of the peripheral retina.52 Microphthalmia and corneal opacities have been observed in families with NDP variants, especially at later stages.51, 53, 54 The variant identified in Patient 3 was previously reported in a patient with Norrie disease, but no details were provided regarding specific ocular or extraocular features.50 The phenotype in Patient 3 and his brother is limited to the eye, which is unusual but has been reported in other patients carrying NDP variants, especially missense changes.50, 53 One study that characterized extraocular features among 56 members (from 39 families) of the Norrie Disease Registry found that missense variants were associated with a later age of onset for hearing loss (75% of individuals with missense variants developed hearing loss by 25 years of age vs 15 years for truncating variants; overall, the age of onset for hearing loss varied from 5 to 48 years old) and a lower incidence of cognitive impairment or behavioral disturbances; however, these associations were not statistically significant. Among 33 participants over age 18 (same as Patient 3) in the registry, only two individuals, both with missense variants, had no extraocular features.50
Interestingly, two out of the three pathogenic variants identified in our study were observed in genes associated with dominant phenotypes despite the seemingly recessive or isolated family history. In one case, the PAX6 variant, parental mosaicism for the variant appears to be responsible for its occurrence in two affected siblings. In the other case, the OTX2 variant, de novo occurrence of a dominant pathogenic allele seems to be most likely but could not be proven due to unavailability of parental samples. The frequency of mosaic and de novo variants appears to be higher than originally expected as evident by several recent publications which identify dominant alleles in families with predicted recessive inheritance.14, 55, 56, 57 In addition, analysis of the final pedigree (NDP) would suggest autosomal recessive inheritance (consanguinity and two siblings as the only affected individuals), but an X-linked recessive pathogenic variant was identified. High genetic heterogeneity, wide phenotypic variability and the ambiguity of inheritance patterns in small pedigrees render WES an efficient method to determine a molecular diagnosis for A/M and coloboma phenotypes. At the same time, prescreening of ‘typical’ cases in known major causative factors represents a valid cost-effective strategy.
References
Williamson KA, FitzPatrick DR : The genetic architecture of microphthalmia, anophthalmia and coloboma. Eur J Med Genet 2014; 57: 369–380.
Skalicky SE, White AJ, Grigg JR et al: Microphthalmia, anophthalmia, and coloboma and associated ocular and systemic features: understanding the spectrum. JAMA Ophthalmol 2013; 131: 1517–1524.
Shah SP, Taylor AE, Sowden JC et al: Anophthalmos, microphthalmos, and typical coloboma in the United Kingdom: a prospective study of incidence and risk. Invest Ophthalmol Vis Sci 2011; 52: 558–564.
Verma AS, Fitzpatrick DR : Anophthalmia and microphthalmia. Orphanet J Rare Dis 2007; 2: 47.
Morrison D, FitzPatrick D, Hanson I et al: National study of microphthalmia, anophthalmia, and coloboma (MAC) in Scotland: investigation of genetic aetiology. J Med Genet 2002; 39: 16–22.
Slavotinek AM : Eye development genes and known syndromes. Mol Genet Metab 2011; 104: 448–456.
Schneider A, Bardakjian T, Reis LM, Tyler RC, Semina EV : Novel SOX2 mutations and genotype-phenotype correlation in anophthalmia and microphthalmia. Am J Med Genet A 2009; 149A: 2706–2715.
Reis LM, Tyler RC, Schneider A et al: FOXE3 plays a significant role in autosomal recessive microphthalmia. Am J Med Genet A 2010; 152A: 582–590.
Schilter KF, Schneider A, Bardakjian T et al: OTX2 microphthalmia syndrome: four novel mutations and delineation of a phenotype. Clin Genet 2011; 79: 158–168.
Gerth-Kahlert C, Williamson K, Ansari M et al: Clinical and mutation analysis of 51 probands with anophthalmia and/or severe microphthalmia from a single center. Mol Genet Genomic Med 2013; 1: 15–31.
Chassaing N, Causse A, Vigouroux A et al: Molecular findings and clinical data in a cohort of 150 patients with anophthalmia/microphthalmia. Clin Genet 2013; 86: 326–334.
Zhu X, Petrovski S, Xie P et al: Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med; e-pub ahead of print 15 January 2015; doi:10.1038/gim.2014.191.
Lopez-Rangel E, Mickelson ECR, Lewis MES : The value of a genetic diagnosis for individuals with intellectual disabilities: optimising healthcare and function across the lifespan. Br J Dev Disabil 2008; 54: 69–82.
Deml B, Reis LM, Maheshwari M, Griffis C, Bick D, Semina EV : Whole exome analysis identifies dominant COL4A1 mutations in patients with complex ocular phenotypes involving microphthalmia. Clin Genet 2014; 86: 475–481.
Deml B, Kariminejad A, Borujerdi RHR, Muheisen S, Reis L, Semina EV : Mutations in MAB21L2 result in Ocular coloboma, microcornea and cataracts. PLOS Genetics 2015; 11: e1005002.
Brady PD, Van Esch H, Fieremans N et al: Expanding the phenotypic spectrum of PORCN variants in two males with syndromic microphthalmia. Eur J Hum Genet 2014; 23: 551–554.
Scott AF, Mohr DW, Kasch LM et al: Identification of an HMGB3 frameshift mutation in a family with an X-linked colobomatous microphthalmia syndrome using whole-genome and X-exome sequencing. JAMA Ophthalmol 2014; 132: 1215–1220.
Reis LM, Tyler RC, Schilter KF et al: BMP4 loss-of-function mutations in developmental eye disorders including SHORT syndrome. Hum Genet 2011; 130: 495–504.
Reis LM, Khan A, Kariminejad A, Ebadi F, Tyler RC, Semina EV : VSX2 mutations in autosomal recessive microphthalmia. Mol Vis 2011; 17: 2527–2532.
Schilter KF, Reis LM, Schneider A et al: Whole-genome copy number variation analysis in anophthalmia and microphthalmia. Clin Genet 2013; 84: 473–481.
Liu X, Jian X, Boerwinkle E : dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat 2013; 34: E2393–E2402.
Richards S, Aziz N, Bale S et al: Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–423.
Xue Y, Ankala A, Wilcox WR, Hegde MR : Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet Med 2014; 17: 444–451.
Hever AM, Williamson KA, van Heyningen V : Developmental malformations of the eye: the role of PAX6, SOX2 and OTX2. Clin Genet 2006; 69: 459–470.
Hanson IM : PAX6 and congenital eye malformations. Pediatr Res 2003; 54: 791–796.
Vincent MC, Pujo AL, Olivier D, Calvas P : Screening for PAX6 gene mutations is consistent with haploinsufficiency as the main mechanism leading to various ocular defects. Eur J Hum Genet 2003; 11: 163–169.
Hanson IM, Fletcher JM, Jordan T et al: Mutations at the PAX6 locus are found in heterogeneous anterior segment malformations including Peters' anomaly. Nat Genet 1994; 6: 168–173.
Dansault A, David G, Schwartz C et al: Three new PAX6 mutations including one causing an unusual ophthalmic phenotype associated with neurodevelopmental abnormalities. Mol Vis 2007; 13: 511–523.
Azuma N, Yamaguchi Y, Handa H, Hayakawa M, Kanai A, Yamada M : Missense mutation in the alternative splice region of the PAX6 gene in eye anomalies. Am J Hum Genet 1999; 65: 656–663.
Azuma N, Yamaguchi Y, Handa H et al: Mutations of the PAX6 gene detected in patients with a variety of optic-nerve malformations. Am J Hum Genet 2003; 72: 1565–1570.
Xiao X, Li S, Zhang Q : Microphthalmia, late onset keratitis, and iris coloboma/aniridia in a family with a novel PAX6 mutation. Ophthalmic Genet 2012; 33: 119–121.
Henderson RA, Williamson K, Cumming S et al: Inherited PAX6, NF1 and OTX2 mutations in a child with microphthalmia and aniridia. Eur J Hum Genet 2007; 15: 898–901.
Solomon BD, Pineda-Alvarez DE, Balog JZ et al: Compound heterozygosity for mutations in PAX6 in a patient with complex brain anomaly, neonatal diabetes mellitus, and microophthalmia. Am J Med Genet A 2009; 149A: 2543–2546.
Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL : PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet 1994; 7: 463–471.
Schmidt-Sidor B, Szymanska K, Williamson K et al: Malformations of the brain in two fetuses with a compound heterozygosity for two PAX6 mutations. Folia Neuropathol 2009; 47: 372–382.
Redeker EJ, de Visser AS, Bergen AA, Mannens MM : Multiplex ligation-dependent probe amplification (MLPA) enhances the molecular diagnosis of aniridia and related disorders. Mol Vis 2008; 14: 836–840.
Park SH, Kim MS, Chae H, Kim Y, Kim M : Molecular analysis of the PAX6 gene for congenital aniridia in the Korean population: identification of four novel mutations. Mol Vis 2012; 18: 488–494.
Zhang X, Tong Y, Xu W et al: Two novel mutations of the PAX6 gene causing different phenotype in a cohort of Chinese patients. Eye (Lond) 2011; 25: 1581–1589.
Chao LY, Huff V, Strong LC, Saunders GF : Mutation in the PAX6 gene in twenty patients with aniridia. Hum Mutat 2000; 15: 332–339.
Thaung C, West K, Clark BJ et al: Novel ENU-induced eye mutations in the mouse: models for human eye disease. Hum Mol Genet 2002; 11: 755–767.
Beby F, Lamonerie T : The homeobox gene Otx2 in development and disease. Exp Eye Res 2013; 111: 9–16.
Ragge NK, Brown AG, Poloschek CM et al: Heterozygous mutations of OTX2 cause severe ocular malformations. Am J Hum Genet 2005; 76: 1008–1022.
Wyatt A, Bakrania P, Bunyan DJ et al: Novel heterozygous OTX2 mutations and whole gene deletions in anophthalmia, microphthalmia and coloboma. Hum Mutat 2008; 29: E278–E283.
Ragge N, Quaghebeur G, Stewart H : SOX2 anophthalmia syndrome in adulthood - a neurodegenerative picture? Clin Genet 2013; 83: 482–484.
Frantz GD, Weimann JM, Levin ME, McConnell SK, Otx1 : and Otx2 define layers and regions in developing cerebral cortex and cerebellum. J Neurosci 1994; 14: 5725–5740.
Vernay B, Koch M, Vaccarino F et al: Otx2 regulates subtype specification and neurogenesis in the midbrain. J Neurosci 2005; 25: 4856–4867.
Larsen KB, Lutterodt MC, Mollgard K, Moller M : Expression of the homeobox genes OTX2 and OTX1 in the early developing human brain. J Histochem Cytochem 2010; 58: 669–678.
de Haas T, Oussoren E, Grajkowska W et al: OTX1 and OTX2 expression correlates with the clinicopathologic classification of medulloblastomas. J Neuropathol Exp Neurol 2006; 65: 176–186.
Bunt J, Hasselt NE, Zwijnenburg DA et al: OTX2 directly activates cell cycle genes and inhibits differentiation in medulloblastoma cells. Int J Cancer 2012; 131: E21–E32.
Smith SE, Mullen TE, Graham D, Sims KB, Rehm HL : Norrie disease: extraocular clinical manifestations in 56 patients. Am J Med Genet A 2012; 158A: 1909–1917.
Sims KB : NDP-Related Retinopathies, in Pagon RA, Adam MP, Ardinger HH et al (eds): GeneReviews (R): Seattle (WA), 1993.
Nikopoulos K, Venselaar H, Collin RW et al: Overview of the mutation spectrum in familial exudative vitreoretinopathy and Norrie disease with identification of 21 novel variants in FZD4, LRP5, and NDP. Hum Mutat 2010; 31: 656–666.
Weh E, Reis LM, Happ HC et al: Whole exome sequence analysis of Peters anomaly. Hum Genet 2014; 133: 1497–1511.
Zhang XY, Jiang WY, Chen LM, Chen SQ : A novel Norrie disease pseudoglioma gene mutation, c.-1_2delAAT, responsible for Norrie disease in a Chinese family. Int J Ophthalmol 2013; 6: 739–743.
Nemirovsky SI, Cordoba M, Zaiat JJ et al: Whole Genome Sequencing Reveals a De Novo SHANK3 Mutation in Familial Autism Spectrum Disorder. PLoS One 2015; 10: e0116358.
Danda S, van Rahden VA, John D et al: Evidence of Germline Mosaicism for a Novel BCOR Mutation in Two Indian Sisters with Oculo-Facio-Cardio-Dental Syndrome. Mol Syndromol 2014; 5: 251–256.
Miyatake S, Koshimizu E, Hayashi YK et al: Deep sequencing detects very-low-grade somatic mosaicism in the unaffected mother of siblings with nemaline myopathy. Neuromuscul Disord 2014; 24: 642–647.
Acknowledgements
We gratefully acknowledge the patients and their families for their participation in research studies. This work was supported by the National Institutes of Health awards R01EY015518 and funds provided by the Children’s Hospital of Wisconsin (EVS), 1UL1RR031973 from the Clinical and Translational Science Award program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Deml, B., Reis, L., Lemyre, E. et al. Novel mutations in PAX6, OTX2 and NDP in anophthalmia, microphthalmia and coloboma. Eur J Hum Genet 24, 535–541 (2016). https://doi.org/10.1038/ejhg.2015.155
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2015.155
This article is cited by
-
ARHGAP35 is a novel factor disrupted in human developmental eye phenotypes
European Journal of Human Genetics (2023)
-
PAX6 missense variants in two families with isolated foveal hypoplasia and nystagmus: evidence of paternal postzygotic mosaicism
European Journal of Human Genetics (2021)
-
Optic nerve coloboma as extension of the phenotype of 22q11.23 duplication syndrome: a case report
BMC Ophthalmology (2020)
-
Genetic analysis of children with congenital ocular anomalies in three ecological regions of Nepal: a phase II of Nepal pediatric ocular diseases study
BMC Medical Genetics (2020)
-
CUGC for syndromic microphthalmia including next-generation sequencing-based approaches
European Journal of Human Genetics (2020)