Abstract
The history of human settlement in Southeast Asia has been complex and involved several distinct dispersal events. Here, we report the analyses of 1825 individuals from Southeast Asia including new genome-wide genotype data for 146 individuals from three Mainland Southeast Asian (Burmese, Malay and Vietnamese) and four Island Southeast Asian (Dusun, Filipino, Kankanaey and Murut) populations. While confirming the presence of previously recognised major ancestry components in the Southeast Asian population structure, we highlight the Kankanaey Igorots from the highlands of the Philippine Mountain Province as likely the closest living representatives of the source population that may have given rise to the Austronesian expansion. This conclusion rests on independent evidence from various analyses of autosomal data and uniparental markers. Given the extensive presence of trade goods, cultural and linguistic evidence of Indian influence in Southeast Asia starting from 2.5 kya, we also detect traces of a South Asian signature in different populations in the region dating to the last couple of thousand years.
Similar content being viewed by others
Introduction
Mainland (MSEA) and Island Southeast Asia (ISEA) are home to hundreds of different ethno-linguistic groups each displaying a complex demographic history.1 Previous studies have revealed strong genetic correlations between populations that are geographically and linguistically close and suggested a common origin of all Southeast Asian and East Asian populations from a single migration wave.2 It is well known, however, that in the more recent past, the populations living in this region have undergone major demographic changes, particularly during the last 5000 years in association with the spread of the Neolithic cultural complex and Austronesian languages.3 Wollstein and colleagues4 reported significant genetic contributions from people currently inhabiting the Borneo (used as a proxy for Asian influence) and Papua New Guinea islands into Malayo-Polynesians (Austronesians who migrated beyond Taiwan) from Near and Remote Oceania. These admixture events were dated to approximately 3 kya, consistently with similar population movements involving people of Asian ancestry moving through ISEA dated around 4–3 kya.5 More recent studies6, 7 have distinguished at least three major ancestral components in MSEA and ISEA in association with Papuan, Austroasiatic- and Austronesian-speaking populations. However, the analyses aiming to identify the likely source regions of these dispersals are confounded by recent admixture in most modern ISEA populations with groups originating from other regions including MSEA2, 8 (see Supplementary Text S1 for more details on the candidate populations included in this study).
In addition to the migratory events involving Southeast Asian sources, more recent South Asian influences in forms of cultural and trading networks, starting more than 2 kya, in ISEA and MSEA have been well established from historical and archaeological data.9, 10, 11, 12
Exemplary for these developments are the sites of Khao Sam Kaeo and Phu Khao Thong from Peninsular Thailand yielding archaeological evidence dating to 2.3–1.2 kya. They confirm the earliest trade networks with India, which include rouletted ware, semi-precious stone beads and artefacts, and Indian crops.13 In ISEA, one finds evidence of Indian trade either directly or via peninsular Thailand. Coastal sites located in Northern Bali dating to 2.1 kya yielded pottery of East Indian or Sri Lankan production, gold and carnelian objects from North India and mung bean.14 Furthermore, epigraphy indicates a strong Indian impact on the nascent political structures of the region15 and provides records of Brahmanic rituals and animal sacrifices.16
Linguistic evidence also supports early interethnic contact between Indian and Southeast Asian populations. Apart from the ubiquitous influence of Sanskrit,17 where it is difficult to distinguish ancient from more recent borrowings, analyses of the earliest Maritime Southeast Asian literature demonstrate that it already exhibits signs of Tamil influence from South India, much of which most likely spread across the region through pre-existing local networks.18 Traces of paternal (Y chromosomes) and maternal (mtDNA) Indian ancestry have been detected across several Indonesian islands at low frequency (<5%).19, 20, 21, 22 The influx of Indian ancestry is detectable in some genome-wide analyses of low density autosomal SNP data2 while being restricted to just a few populations from western Indonesia (Sumatra). Contrarily to that, a more recent study23 using medium density SNP data could not find a South Asian genetic signature in Southeast Asia. The same authors, however, inferred gene flow from the Indian subcontinent to Aboriginal Australian populations and dated it at around 4 kya. In the absence of a similar South Asian component in SEA, this finding was interpreted to require a direct sea route bypassing Southeast Asia to explain such a signature in Australasia.
In order to refine the current understanding on the source of the Austronesian expansion and to further explore potential South Asian genetic contributions in MSEA and ISEA, we generated high density (730K) SNP Chip data for a panel of 196 individuals from 10 populations including 50 of which (from the Bajo and Lebbo populations) are published already7 and 146 new (Burmese and Vietnamese from MSEA; Ilocano, Tagalog and Kankanaey from the Philippines; Murut, Malay and Dusun from ISEA plus four Australian Aborigines). We merged the newly generated dataset with those available from the literature (cf. Material and Methods) and (i) investigated the existence of signs of South Asian admixture in our new SEA populations, (ii) refined current knowledge on the putative source of the Austronesian expansion; (iii) determined the extent to which signs of local adaptation are shared across local populations, as function of their common demographic history.
Materials and methods
Samples, genotyping and phasing
The newly generated dataset for this study consists of 150 individuals from nine Southeast Asian and one Australian population (Figure 1 and Supplementary Table S10). DNA was extracted from saliva samples collected from healthy adult donors who signed an informed consent form. The study was approved by local Research Ethics Committees (SingHealth Centralised Institutional Review Board and the Medical and Health Research Ethics Committee of the National Cancer Centre, Brunei Darussalam), the Cambridge Ethics Committee (HBREC.2011.01) and the ERC Ethics Panel. Southeast Asian samples were genotyped using Illumina (San Diego, CA, USA) OmniExpress Bead Chips for 730 525 SNPs. They are accessible together with 50 Bajo and Lebbo samples under the GEO accession number GSE77508. For the three Australian samples, the Illumina Human 660K Quad Bead Chip yielded 655 215 SNPs, while for one Australian, the 610K version of the latter chip gave 616 795 variants. These four samples are accessible under the accession number EGAS00001001738 in the European Genome-phenome Archive.
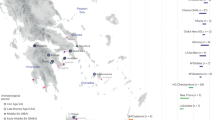
(a) A map of Southeast Asia, displaying a subset of populations assessed in this study and the distribution of ancestry components based on the local ADMIXTURE run with the optimal number of ancestry components (K=9, cf. Supplementary Figure S2). The figure legend on the lower left section shows the list of genetic ancestry components whose colour codes correspond to those on the pie charts. Components k8 and k9 are mainly present in the Yoruba and Ati Negritos, respectively, and do not significantly contribute to the genetic diversity of the groups displayed in Figure 1. The population abbreviations are as follows: Alo-Alorese, Baj-Bajo, Bat-Batak, Bru-Brunei (Dusun, Murut), Bur- Burmese, CHB-Chinese from Beijing, Jav-Javanese, Kan-Kankanaey Igorots, Leb-Lebbo, Mal-Malay, Mam-Mamanwa Negritos, Men-Mentawai, Mun-Mundari, NIn-North Indians, Pap-Papuans, PhU-Philippine Urban, SIn-South Indians, Taw-Ami and Atayal from Taiwan, Viet-Vietnamese. Note that the symbols next to the population names reflect the linguistic affiliations. Austroasiatic languages: circle, Austronesian languages: asterisk, Indo-European languages: square, Dravidian languages: hash, Papuan languages: cross, Tibeto-Burman languages: caret. (b) Three graphs showing the proportions of ancestry components k3, k4 and k6 from their emergence as independent components in the Papuans (k3, red), Indian populations (k4, green) and the Kankanaey Igorot (k6, brown) across multiple higher K values. All populations displayed show a percentage of at least 5% of the respective ancestry when it emerges.
Before the analyses as such, data filtering and quality checks using PLINK 1.0724 were performed. First, only autosomal SNPs with a genotyping success rate greater than 98% were included. PLINK was also utilised to remove all individuals more closely related than first degree cousins. This was carried out by estimating pairwise identity by descent iteratively; individuals with an identity by descent>0.125 were excluded. Following these quality controls, haplotypes were inferred from genotype data with SHAPEIT.25
Furthermore, eight full mitochondrial Kankanaey genomes were sequenced by Complete Genomics (Mountain View, CA, USA) using CG software version 2.4. Access to the sequences is provided under the GenBank accession numbers KU752558 to KU752565. All novel data from this paper will also be available under www.ebc.ee/free_data in the PLINK (genotype data) and fasta (mitochondrial genomes) formats, respectively.
Demographic analyses
To get a first overview for the novel Southeast Asian data, we merged them with four reference populations from the HapMap 3 panel26 and the HGDP Papuans27 to obtain a set of 307 625 common SNPs. Runs of homozygosity, average observed heterozygosity and identity by descent were obtained using PLINK default parameters. Furthermore pairwise FST was calculated using an ad hoc Perl script implementing an estimator for Wright’s formula.28
To address more specific questions regarding the ancestries of our novel populations, we performed two distinct ADMIXTURE29, 30 analyses. For comparative purposes, publicly available genotype data from the HapMap,26 HDGP27 and the Pan-Asian Consortium2 projects were added to 185 individuals from nine SEA populations (the divergence from the original number of 196 is because of the removal of close relatives). In addition, we used SNP data from studies focused on Indian populations.31, 32 This resulted in a dataset consisting of 1099 individuals.
For further verification of our ADMIXTURE analysis, we assembled a second panel of 1010 samples including 187 samples from our nine SEA populations, and four Australian Aborigines, which are newly reported here. The samples, populations and references for both analyses are listed in Supplementary Table S10. A detailed description of the merging and data curation for ADMIXTURE can be found in the Supplementary Text S2.
Effective population size for our nine SEA populations was estimated by analysing linkage disequilibrium patterns with the NeON R package.33 To further investigate genetic structure and gene flow between populations we used the TreeMix v1.1 software package.34 To measure how well the trees with different numbers of migration events (N) reflect the relationship between population groups, we calculated the fraction f of explained variance as described by the original authors of the method. We used MEGA v6.0.635 to create a graphic representation of the TreeMix output. For specific admixture events of interest suggested by the ADMIXTURE plots, the respective sets of recipient and source populations were tested with the three populations test (f3).34, 36 The population trios yielding a Z-score smaller than −2 were considered significantly admixed. These were then analysed with ALDER37 to date the putative admixture event. Furthermore, we used the f4-ratio test38 to obtain a quantitative estimate of admixture percentages of interest.
For the analysis of the mtDNA data, the haplogroup affiliation of each sample was assigned using HaploGrep 2.039 and PhyloTree build 16 (as of 19/02/2014) (http://www.phylotree.org).40 The variants are described relative to the rCRS (GenBank Accession Number NC_012920.1).41
Selection tests
To capture haplotype homozygosity-based signals, the Integrated Haplotype Score (iHS)42 and Cross Population Extended Haplotype Homozygosity (XP-EHH)43 tests were used. Both the iHS and XP-EHH statistics were calculated as in the study by Pickrell et al.,44 yielding about 10 000–11 000 genomic windows for iHS and about 13 700 windows for XP-EHH for each SEA population analysed. From the top 1% of all iHS signals, putatively the strongest candidates for selection, windows present in the top 5% iHS windows of the CHB population from the HapMap panel were excluded, to pick up only signals particular to SEA. However, for the analysis of regional sharing patterns based on the iHS, this condition did not apply. For the XP-EHH, the use of a reference population is inherent in the method; again CHB was chosen, for similar reasons.
Furthermore, we computed the allele frequency-based Population Branch Statistic. This test statistic represents the amount of allele frequency change at a given locus in the history of the test population since it diverged from other populations.45 The outgroups for each tested SEA population were the YRI and CHB populations. Pairwise FST values for the populations of interest and the references were calculated following Weir and Cockerham.46 Population Branch Statistic scores were estimated from the pairwise FST values.45 On the basis of the approach of Pickrell et al.,44 the genome was divided into windows of a modified size of 100 kb and the maximum Population Branch Statistic score in each window was used as the test statistic. This resulted in between 26 000 and 27 000 windows for each analysed group.
Results
To investigate general patterns of population structure in our data, we performed two distinct ADMIXTURE analyses: the first was mainly focused on populations from Southeast Asia and South Asia, while the second provided the context of a broader, worldwide genetic landscape and additional validation for inferences from the first analysis.
According to the cross-validation scores for both analyses, K=9 admixture fractions provide the best fit (for the local plot, additional Ks are provided in Supplementary Figure S2, for the global plot, Ks from 3 to 15 are shown in Supplementary Figure S3B). The ADMIXTURE analyses of the newly generated data (Figure 1a, Supplementary Figure S1) recapitulate the main ancestral components associated with Austronesian (k6), Austroasiatic (k5) and Papuan (k3) populations (Figure 1, Supplementary Figure S2) already described in the area by previous studies.5, 6 At lower K values, the component associated with the Papuans is highly prevalent in Eastern Indonesia and the Mamanwa (a Negrito group from the Philippines), while at higher values, it continues to persist only in the Alorese and Bajo from Indonesia (Figure 1b, Supplementary Figure S2).
Burmese and Vietnamese exhibit significant proportions of the k2 component indicating shared ancestry with East Asian populations. The k4 component associated with South Asian ancestry is also consistently visible in Burmese and Malays (this study) and some Indonesian populations, mainly the Batak of Sumatra.2 However, at lower Ks, this component is also present in the Javanese and the Mamanwa Negritos, suggesting affinities that, however, decline with higher Ks (Figure 1b, Supplementary Figure S2). Notably, in the extended worldwide analysis (Supplementary Figure S3B), the Papuan-related component (red) in the Bajo and the South Asian signal (green) in the Burmese and Malays were also clearly detectable. The SEA groups described here exhibit a remarkable diversity from very heterogeneous groups such as the Malays to the Kakanaey who appear homogenous in their ancestry composition by the ADMIXTURE analyses (Figure 1b, Supplementary Figure S2).
The Kankanaey are an indigenous population of northern Luzon, belonging to the broader ‘Igorot’ group. At K=9, the majority of Kankanaey ancestry is in the k6 component, which they share with the Ami (AX-AM) and Atayal (AX-AT) from Taiwan and, hence, is putatively associated with the Austronesian expansion (Figure 1a, Supplementary Figure S2). When it emerges as distinct from the other Asian components, the k6 brown ancestry is spread throughout ISEA and remains stable in all these groups from K8-10 (Figure 1b, Supplementary Figure S2). Remarkably, in the regional admixture plots, the Kankanaey remain unadmixed throughout all Ks from 2 to 10 (Supplementary Figure S2), even though at lower Ks, they do not yet have their own distinct component. These findings are consistent with the Kankanaey’s geographic location, the Mountain Province in the Northern Philippines (Figure 1a, Supplementary Figure S1), close to Taiwan, the likely centre of the Austronesian expansion.3, 6
Kankanaey genome-wide heterozygosity levels and extent of runs of homozygosity (Supplementary Table S1) rule out potential confounders such as extreme inbreeding or genetic drift being causative for their unusually homogeneous ancestry. To further explore the potential effect of demographic history on population structure, we estimated the effective population size of the nine SEA populations presented here based on the development of linkage disequilibrium patterns over time (Supplementary Figure S4).33 The mainland Burmese and Vietnamese groups exhibit comparatively high effective population sizes and signs of recent expansion. This is in line with their recent history of admixture with neighbouring populations, whereas there is more variation in the ISEA populations. Notably, the Kankanaey have one of the lowest values varying between 2000 and 3000 (6000–27 000 kya). However, they are not an extreme outlier and are comparable with the Lebbo from Borneo (no significant difference, P=0.7938), who instead do not show such a homogeneous ADMIXTURE profile. Under the assumption that the brown k6 component reflects ancestry connected to the Austronesian expansion, the Kankanaey displayed a higher percentage of it than even Austronesian Taiwanese populations (AX-AT, AX-AM, Figure 1a, Supplementary Figure S2). The affinity of the Kankanaey to these groups was supported by the TreeMix34 analyses of 25 populations (Supplementary Figures S5 and S6) where the Kankanaey did not cluster with other Filipinos but rather with the Taiwanese aboriginals.
The emerging picture seems to be compatible with a scenario of local Austroasiatic and Papuan components influenced by the incoming Austronesian (brown k6, Figure 1a, Supplementary Figure S2) wave 4–3 kya, which originated from a population living in Taiwan and, perhaps, in the North Philippines.6 The attempt to date the above admixture events using ALDER37 highlighted a clear admixture pattern between ‘Kankanaey like’ people and earlier substrates, dated to at least 2.2 kya in the Bajo (Table 1).
These affinities of the Kankanaey and their potential role as a good proxy for the Austronesian expansion are further highlighted when looking at uniparental markers. The eight available Kankanaey mtDNA sequences (Supplementary Table S2) exhibit lineages (B4a1a;M7b1a2a1) that are typical markers of Malayo-Polynesian-speaking populations.47, 48
Finally, the Kankanaey cannot be modelled as any kind of mixture from 46 populations using the f3 statistic (Supplementary Table S3).36 Taken together, the evidence from these independent approaches suggests that the Kankanaey could potentially represent an unadmixed remnant population close to the source that may have given rise to the Austronesian expansion.
We also utilised the f3 test together with ALDER to further contextualise the potential South Asian connections of some SEA groups. Both of these statistics (Table 1) suggest the presence of variable degrees of South Asian-related ancestry in the MSEA and ISEA populations (Bajo, Burmese, Filipino and Malay). Assuming a generation time of 30 years,49 the earliest possible midpoint of the South Asian admixture is estimated at 2.4 kya. The overall proportion of South Asian ancestry was further estimated by applying the f4 statistic38 (Supplementary Table S4) according to the tree presented in Supplementary Figure S7. The estimated values were 24.9% for the Burmese, 8.3% for the Malays and 5.3% for the Bajo. One limitation of this approach is its dependence on shared genetic drift. As the Papuans and South Indians have a similar position in the phylogenetic tree relative to the other groups, Papuan ancestry could be mistaken as South Indian. This has probably no effect in the Burmese and Malay, who do not show Papuan admixture (Figure 1a, Supplementary Figure S2) but could contribute to the South Indian ancestry detected in the Bajo. True Indian ancestry in this population still seems conceivable given the presence of South Asian lineages in uniparental marker analyses.22
These analyses indicate a South Asian-related component in the genetic make-up of at least some SEA groups that entered their gene pool ca. 2.4 kya ago, being supported by ADMIXTURE, f3 and f4 analyses for the Burmese and the Malay and by f-statistics for the Bajo (f3, f4) and the lowland Filipinos (f3).
As an additional tool to explore relationships among populations, we examined patterns of haplotype homozygosity and allelic differentiation using test statistics iHS,42 XP-EHH43 and Population Branch Statistic test45 (Supplementary Tables S7). For the iHS, the amount of signal sharing between two groups correlates only very weakly (r2=0.041 for a linear regression) to overall genetic similarity as expressed by the FST (Supplementary Figure S8). However, the MSEA groups and the Han Chinese (included as a reference) who share a considerable proportion of East Asian ancestry (Figure 1a, Supplementary Figure S2) also show a great affinity to each other regarding haplotype homozygosity patterns (Supplementary Table S5). In ISEA, those groups with at least three significant ancestry components at K=9 (Bajo, Filipino, Malay, Figure 1a) exhibit more signal sharing. In contrast, Kakanaey, Lebbo and Murut show reduced sharing with all other populations, which perhaps highlights the phenomena of deep population splits and separate demographic histories in recent times when the haplotype homozygosities have accumulated.
However, these inferences are highly dependent upon the approach utilised. A different picture presents itself for the XP-EHH, which considers both haplotype homozygosity and allelic differentiation, with the Han Chinese used as outgroup. The average fraction of signal sharing declines from 0.31 to 0.22, while the correlation with the FST increases considerably (r2=0.256). This is probably because signals connected to shared ancestry with East Asians are excluded. It causes the Burmese, who exhibit a large fraction of the k2 East Asian-related component (Figure 1a, Supplementary Figure S2) to become an outlier especially with respect to their high fraction of unique top 1% XP-EHH signals, only 15% of which are shared with other groups on average.
Discussion
In this study, we set out to explore the population structure in MSEA and ISEA and more specifically, to clarify the exact nature of South Asian gene flow into SEA and the presence of potential unadmixed Austronesian population(s) close to the ancestral Austronesian source.
We detected a minor South Asian component in our ADMIXTURE analyses in MSEA and ISEA populations (green k4, Figure 1a, Supplementary Figure S2; green, Supplementary Figure S3B), which was further confirmed by f3, f4 and ALDER results and dated to have entered SEA from 2.4kya (Table 1). Although this component is more widespread at lower Ks (Figure 1b, Supplementary Figure S2), at the best K=9 (Figure 1a), the evidence is strongest for the Burmese and the Malay and somewhat weaker for Bajo and Filipinos, where it is limited to the f-statistics (Table 1, Supplementary Table S4). It is important to explore how these results relate to the linguistic and archaeological evidences, attesting a continuous presence of South Asian cultures in Southeast Asia since 2.5 kya.12, 17, 50, 51 This should be performed keeping in mind that in the majority of SEA populations, the Indian component is absent or below the scale of a potential error and detectability. First, it is most likely that the ‘carriers’ of South Asian culture were traders, artisans50 and at a later date, religious scholars (Brahmins) who were influential as advisers to Southeast Asian rulers. Some of these might have been locals educated in India who brought home Sanskrit texts and Brahmanic rituals.52 Therefore, this rather small group would not have left a major genetic signature. Second, the epigraphic record and evidence from monumental archaeology during the late first millennium CE attests that the Indian presence is biased towards courts and generally higher social strata, which can lead us to overestimate the impact on the majority of the population.52 More generally speaking, there are a wide range of scenarios relating to the spread of cultural elements and gene flow and the patterns of this relationship are highly complex to model (cf. the example of the Neolithization in Europe53). Therefore, with the exception of the Burmese, who are also geographically very close to the Indian subcontinent, the evidence points to rather minor Indian gene flow, in contrast to the documented cultural influence which, however, overlaps with the admixture range dated with ALDER (Table 1). This low South Asian gene flow was, however, also detected in some other populations across ISEA.2, 19, 20, 21, 22 Taken together, these findings suggest Southeast Asia as a potential waypoint for the reported South Asian migration into Australasia, which was disputed by the authors who proposed this migration event.23 However, the date obtained using ALDER (2.4 kya) is at least 1500 years posterior to the reported South Indian migration into Australasia.23 A preliminary conclusion would envisage the SEA and Australasia migrations as two separate events. Besides the fact that the dating methods were different in our case and Pugach et al.23 (they used a method based on wavelet transform analysis), at least two caveats can be brought up to reconcile this fragmented scenario. Given the evidence presented here, it seems reasonable to assume a constant gene flow from South Asia into SEA via land, with Australasia being only a sporadic end point. In this case, the 4 kya estimate provided by Pugach and colleagues would be a point estimate of the sparse arrival into Australasia, while our ALDER estimate should be interpreted as the midpoint37 of such a flow between 4 kya and more recent times. Second, given the surprising concordance of linguistic and archaeological evidences for a South Asian presence in SEA around 2.5 kya, one could imagine a particularly intense corresponding gene flow during that time further biasing the ALDER estimate toward this period.
In this study, we have identified the Kankanaey from the northern Philippines as the population harbouring the highest reported amount of the Austronesian genomic component, even higher than the ones detectable in modern aboriginal Taiwanese (Figure 1b, Supplementary Figure S2). This conclusion rests on evidence from several independent analyses including ADMIXTURE, f3, runs of homozygosity, TreeMix, Ne and uniparental markers.
The Kankanaey belong to the broader group of populations collectively known as Igorot (Supplementary Text S1). Various studies exist on the Kakanaey language54 and customs,55 although works on their prehistory are lacking. Genetic data from 30 Kankanaey speakers were included in a recent study of the mtDNA-haplotype diversity in the Philippines.56 There they were shown to share many lineages with two other Igorot groups (Ibaloi and Ifugao) from Northern Luzon. These results are broadly consistent with the uniparental data we present here (Supplementary Table S2), where the Kankanaey show haplotypes also found in Taiwanese aboriginals57 and generally associated with the Austronesian expansion.47, 48 We conclude that the Kankanaey are either the best preserved source of the Austronesian expansion or a case of total replacement that followed it. The dominant model suggests a southward diffusion of Austronesians from Taiwan around 4000 BP, which impacted the Philippines, the north of Borneo and Sulawesi between 3800 and 3600 BP, and later spread into the Pacific.3 Even if the modality of this expansion is complex and still debated,58 the location of the Kankanaey in the northern Philippines, close to Taiwan, suggests that they may be considered as one of the least admixed living groups tracing their ancestry from the source populations of the Austronesian expansion. Furthermore, we confirm the finding of an Austroasiatic-related component in ISEA populations (here the Dusun, Murut, Lebbo and Bajo) first reported by Lipson et al.6 and there described as unexpected owing to the historically nearly exclusive presence of Austroasiatic speakers on the mainland. Given its wide spread in MSEA and ISEA in linguistically diverse groups, the explicit association of k5 with this language family should be taken with caution. However, it is worthwhile noting that in India, we find this component specifically in Munda-speaking populations. The k5 component could represent an ancestral substrate, which was once distributed widely throughout SEA and was encountered by the Austronesians when they spread from Taiwan. Another possibility is that there was an early split into several subgroups during the Austronesian expansion and that this component belongs to the ancestral make-up of a subgroup of Malayo-Polynesians who expanded into western Indonesia.
Our comparison of haplotype-based scans of positive selection revealed that compared with earlier studies on a continental level32 in a regional context in ISEA, there is no good correlation between haplotype sharing patterns and genetic distance as indicated by the FST (Supplementary Figure S8). However, as described above, the haplotype homozygosity patterns still reflect demography to a considerable extent. Populations showing more diversity in the admixture plots also exhibit higher levels of shared signals with other groups. Furthermore, the sharing patterns proved to be very dependent on the kind of test utilised. Notably when the XP-EHH, which uses the Han Chinese as outgroup, is applied, all signals shared with East Asians are excluded. Intriguingly, this causes the Burmese whose ancestry contains a significant South Asian-related component (Figure 1a, Supplementary Figure S2) to become an outlier (Supplementary Table S6) potentially reflecting haplotype homozygosity signals unique to their share of Indian ancestry.
In conclusion, we report a minor South Asian contribution to the genomes of some modern MSEA and ISEA populations, mainly the Burmese and the Malay. This is in line with a general cultural diffusion process to SEA, driven by smaller groups of influential individuals from South Asia. Secondly, our work strongly suggests that based on the currently available data, the Kankanaey tribal group from Northern Luzon, Philippines are the best genetic representative of the Austronesian expansion. We envisage high coverage whole genome sequencing of this population as a sound approach to further explore this major peopling event that shaped the genetic landscape of the broader Southeast Asia region.
Accession codes
References
Lewis MP, Simons GF, Fennig CD (eds): Ethnologue: Languages of the World. 18th edn. SIL International: Dallas, TX, USA, 2015.
HUGO Pan-Asian SNP, Consortium, Abdulla MA, Ahmed I, Assawamakin A et al: Mapping human genetic diversity in Asia. Science 2009; 326: 1541–1545.
Bellwood PS : Prehistory of the Indo-Malaysian Archipelago. ANU E Press: Canberra, 2007.
Wollstein A, Lao O, Becker C et al: Demographic history of Oceania inferred from genome-wide data. Curr Biol 2010; 20: 1983–1992.
Xu S, Pugach I, Stoneking M, Kayser M, Jin L : Genetic dating indicates that the Asian–Papuan admixture through Eastern Indonesia corresponds to the Austronesian expansion. Proc Natl Acad Sci USA 2012; 109: 4574–4579.
Lipson M, Loh P-R, Patterson N et al: Reconstructing Austronesian population history in Island Southeast Asia. Nat Commun 2014; 5: 4689.
Pierron D, Razafindrazaka H, Pagani L et al: Genome-wide evidence of Austronesian-Bantu admixture and cultural reversion in a hunter-gatherer group of Madagascar. Proc Natl Acad Sci USA 2014; 111: 936–941.
Trejaut JA, Poloni ES, Yen J-C et al: Taiwan Y-chromosomal DNA variation and its relationship with Island Southeast Asia. BMC Genet 2014; 15: 77.
Ardika W, Bellwood PS : Sembiran: the beginnings of Indian contact with Bali. Antiquity 1991; 65: 221–232.
Ardika W, Bellwood PS, Sutaba IM, Yuliati KC : Sembiran and the first Indian contacts with Bali: an update. Antiquity 1997; 71: 193–195.
Lawler A : Sailing Sinbad’s seas. Science 2014; 344: 1440–1445.
Manguin P-Y, Mani A, Wade G (eds): Early Interactions Between South and Southeast Asia: Reflections on Cross-cultural Exchange. Institute of Southeast Asian Studies: Singapore; Manohar India, New Delhi, 2011.
Castillo C : The Archaeobotany of Khao Sam Kaeo and Phu Khao Thong: The Agriculture of Late Prehistoric Southern Thailand. Doctoral thesis, University College London, 2013.
Calo A, Prasetyo B, Bellwood P et al: Sembiran and Pacung on the north coast of Bali: a strategic crossroads for early trans-Asiatic exchange. Antiquity 2015; 89: 378–396.
Mabbett IW : The ‘Indianization’ of Southeast Asia: Reflections on the Historical Sources. J Southeast Asian Stud 1977; 8: 143–161.
Guy J : Tamil merchants and the Hindu-Buddhist Diaspora in early Southeast Asia. In: Manguin P-Y, Mani A, Wade G (eds): Early Interactions Between South and Southeast Asia: Reflections on Cross-cultural Exchange. Institute of Southeast Asian Studies: Singapore; Manohar India: New Delhi 2011, pp 243–262.
Gonda J : Sanskrit in Indonesia. International Academy of Indian Culture: New Delhi, 1973.
Hoogervorst T : Detecting pre-modern lexical influence from South India in Maritime Southeast Asia. Archipel 2015; 89: 63–93.
Chaubey G, Endicott P : The Andaman Islanders in a regional genetic context: reexamining the evidence for an early peopling of the archipelago from South Asia. Hum Biol 2013; 85: 153–172.
Karafet TM, Lansing JS, Redd AJ et al: Balinese Y-chromosome perspective on the peopling of Indonesia: genetic contributions from pre-neolithic hunter-gatherers, Austronesian farmers, and Indian traders. Hum Biol 2005; 77: 93–114.
Karafet TM, Hallmark B, Cox MP et al: Major east-west division underlies Y chromosome stratification across Indonesia. Mol Biol Evol 2010; 27: 1833–1844.
Kusuma P, Cox MP, Brucato N, Sudoyo H, Letellier T, Ricaut F-X : Western Eurasian genetic influences in the Indonesian archipelago. Quat Int 2015, e-pub ahead of print 18 August 2015; doi:10.1016/j.quaint.2015.06.048.
Pugach I, Delfin F, Gunnarsdóttir E, Kayser M, Stoneking M : Genome-wide data substantiate Holocene gene flow from India to Australia. Proc Natl Acad Sci USA 2013; 110: 1803–1808.
Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Delaneau O, Zagury J-F, Marchini J : Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods 2013; 10: 5–6.
International HapMap Consortium, Frazer KA, Ballinger DG, Cox DR et al: A second generation human haplotype map of over 3.1 million SNPs. Nature 2007; 449: 851–861.
Li JZ, Absher DM, Tang H et al: Worldwide human relationships inferred from genome-wide patterns of variation. Science 2008; 319: 1100–1104.
Wright S : Evolution in Mendelian populations. Genetics 1931; 16: 97–159.
Alexander DH, Novembre J, Lange K : Fast model-based estimation of ancestry in unrelated individuals. Genome Res 2009; 19: 1655–1664.
Cardona A, Pagani L, Antao T et al: Genome-wide analysis of cold adaptation in indigenous Siberian populations. PLoS One 2014; 9: e98076.
Chaubey G, Metspalu M, Choi Y et al: Population genetic structure in Indian Austroasiatic speakers: the role of landscape barriers and sex-specific admixture. Mol Biol Evol 2011; 28: 1013–1024.
Metspalu M, Romero IG, Yunusbayev B et al: Shared and unique components of human population structure and genome-wide signals of positive selection in South Asia. Am J Hum Genet 2011; 89: 731–744.
Mezzavilla M, Ghirotto S : Neon: An R package to estimate human effective population size and divergence time from patterns of linkage disequilibrium between SNPS. J Comput Sci Syst Biol 2015; 8: 037–044.
Pickrell JK, Pritchard JK : Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet 2012; 8: e1002967.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S : MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30: 2725–2729.
Reich D, Thangaraj K, Patterson N, Price AL, Singh L : Reconstructing Indian population history. Nature 2009; 461: 489–494.
Loh P-R, Lipson M, Patterson N et al: Inferring admixture histories of human populations using linkage disequilibrium. Genetics 2013; 193: 1233–1254.
Moorjani P, Patterson N, Hirschhorn JN et al: The history of African gene flow into Southern Europeans, Levantines, and Jews. PLoS Genet 2011; 7: e1001373.
Kloss-Brandstätter A, Pacher D, Schönherr S et al: HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum Mutat 2011; 32: 25–32.
van Oven M, Kayser M : Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 2009; 30: E386–E394.
Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N : Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 1999; 23: 147.
Voight BF, Kudaravalli S, Wen X, Pritchard JK : A map of recent positive selection in the human genome. PLoS Biol 2006; 4: e72.
Sabeti PC, Varilly P, Fry B et al: Genome-wide detection and characterization of positive selection in human populations. Nature 2007; 449: 913–918.
Pickrell JK, Coop G, Novembre J et al: Signals of recent positive selection in a worldwide sample of human populations. Genome Res 2009; 19: 826–837.
Yi X, Liang Y, Huerta-Sanchez E et al: Sequencing of 50 human exomes reveals adaptation to high altitude. Science 2010; 329: 75–78.
Weir BS, Cockerham CC : Estimating F-statistics for the analysis of population structure. Evolution 1984; 38: 1358–1370.
Trejaut JA, Kivisild T, Loo JH et al: Traces of archaic mitochondrial lineages persist in Austronesian-speaking Formosan populations. PLoS Biol 2005; 3: e247.
Soares P, Rito T, Trejaut J et al: Ancient voyaging and Polynesian origins. Am J Hum Genet 2011; 88: 239–247.
Fenner JN : Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol 2005; 128: 415–423.
Bellina B, Silapanth P, Chaisuwan B et al. The development of coastal polities in the upper Thai-Malay Peninsula in the late first millennium BCE. In: Revire N, Murphy SA(eds): Before Siam: Essays in Art and Archaeology. River Books: Bangkok, 2014, pp 69–89..
Calo A : Ancient trade between India and Indonesia. Science 2014; 345: 1255–1255.
Bronkhorst J : The spread of Sanskrit in Southeast Asia. In: Manguin P-Y, Mani A, Wade G (eds): Early Interactions Between South and Southeast Asia: Reflections on Cross-Cultural Exchange. Institute of Southeast Asian Studies: Singapore; Manohar India: New Delhi 2011, pp 263–275.
Fort J : Demic and cultural diffusion propagated the Neolithic transition across different regions of Europe. J R Soc Interface 2015; 12: 20150166–20150166.
Allen JL : Kankanaey: a Role and Reference Grammar Analysis. SIL International Publications: Dallas, Texas, USA, 2014.
Kohnen N : ‘Natural’ childbirth among the Kankanaly-Igorot. Bull N Y Acad Med 1986; 62: 768–777.
Delfin F, Min-Shan Ko A, Li M et al: Complete mtDNA genomes of Filipino ethnolinguistic groups: a melting pot of recent and ancient lineages in the Asia-Pacific region. Eur J Hum Genet 2014; 22: 228–237.
Ko AM-S, Fu Q, Chen CY et al: Early Austronesians: into and out of Taiwan. Am J Hum Genet 2014; 94: 426–436.
Bulbeck F : An integrated perspective on the Austronesian Diaspora: the switch from cereal agriculture to maritime foraging in the colonisation of Island Southeast Asia. Aust Archaeol 2008; 67: 31–52.
Acknowledgements
This work was supported by the European Research Council Starting Investigator grant FP7-261213 to TK. This work was supported by the French ANR grant number ANR-14-CE31-0013-01 (grant OceoAdapto to F-XR), the French ANR-12-PDOC-0037-01 (grant GENOMIX to DP), the Region Aquitaine of France (grant MAGE to TL), the French Ministry of Foreign and European Affairs (French Archaeological Mission in Borneo (MAFBO) to F-XR). TA was supported by a Wellcome Trust Post-Doctoral fellowship (WT1000MA).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Mörseburg, A., Pagani, L., Ricaut, FX. et al. Multi-layered population structure in Island Southeast Asians. Eur J Hum Genet 24, 1605–1611 (2016). https://doi.org/10.1038/ejhg.2016.60
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2016.60
This article is cited by
-
Reanalyzing the genetic history of Kra-Dai speakers from Thailand and new insights into their genetic interactions beyond Mainland Southeast Asia
Scientific Reports (2023)
-
Patterns of African and Asian admixture in the Afrikaner population of South Africa
BMC Biology (2020)
-
The Early Peopling of the Philippines based on mtDNA
Scientific Reports (2020)
-
The genetic legacy of continental scale admixture in Indian Austroasiatic speakers
Scientific Reports (2019)
-
Investigating the origins of eastern Polynesians using genome-wide data from the Leeward Society Isles
Scientific Reports (2018)