Abstract
The peopling of Greenland has a complex history shaped by population migrations, isolation and genetic drift. The Greenlanders present a genetic heritage with components of European and Inuit groups; previous studies using uniparentally inherited markers in Greenlanders have reported evidence of a sex-biased, admixed genetic background. This work further explores the genetics of the Greenlanders by analysing autosomal and X-chromosomal data to obtain deeper insights into the factors that shaped the genetic diversity in Greenlanders. Fourteen Greenlandic subsamples from multiple geographical settlements were compared to assess the level of genetic substructure in the Greenlandic population. The results showed low levels of genetic diversity in all sets of the genetic markers studied, together with an increased number of X-chromosomal loci in linkage disequilibrium in relation to the Danish population. In the broader context of worldwide populations, Greenlanders are remarkably different from most populations, but they are genetically closer to some Inuit groups from Alaska. Admixture analyses identified an Inuit component in the Greenlandic population of approximately 80%. The sub-populations of Ammassalik and Nanortalik are the least diverse, presenting the lowest levels of European admixture. Isolation-by-distance analyses showed that only 16% of the genetic substructure of Greenlanders is most likely to be explained by geographic barriers. We suggest that genetic drift and a differentiated settlement history around the island explain most of the genetic substructure of the population in Greenland.
Similar content being viewed by others
Introduction
The peopling of Greenland was the result of a complex set of population migrations and adaptation to the harsh living conditions of the arctic setting. The earliest inhabitants of Greenland were Paleo-Eskimos, comprising distinct extinct groups that are archaeologically associated with (1) the Independence I – Saqqaq and Pre-Dorset cultures, present approximately 3900 to 2500 years ago (YA), and (2) the Independence II – Dorset culture, which lasted from 2500 until 700 YA.1 Approximately 1500 YA, Neo-Eskimo coming from Alaska – the Thule – spread to the East and settled in Greenland.2, 3, 4 In 985 AD, the Norse, who arrived from Iceland, settled on the East coast of Greenland and founded Norse communities that existed for approximately 500 years.5 In the eighteenth century, another wave of immigration reached Greenland, mostly from Norway and Denmark.5
As of 2009, Greenland has four municipalities: Kujalleq, Semersooq, Qeqqata and Qaasuitsup (Figure 1). The number of inhabitants per municipality ranges from 7151 in Kujalleq to 21 868 in Semersooq, with a total of 56 370 inhabitants on the entire island (http://www.stat.gl; data from January 2013). Owing to the harsh climate, the majority of the population is concentrated on the West coast.
Studies based on uniparentally inherited markers have shown that the Y-chromosomal gene pool of Greenlandic people comprises approximately equal numbers of European and Inuit lineages,6, 7, 8, 9 but the set of mtDNA haplogroups revealed an overwhelmingly Inuit component, with no European lineages detected.10 More recently, Helgason et al4 reported evidence of an intricate pattern of mtDNA variation in Greenlanders. The analysis of different regions within Greenland showed high heterogeneity on the island, suggesting that, in addition to the Thule, other Inuit might have contributed to the current-day genetic variation of Greenlanders. The complexity of the Greenlandic population was also affirmed through recent studies based on whole-genome and mtDNA analyses of a Paleo-Eskimo.1, 11 Taken together, these studies provided indications of a sex-biased and heterogeneous process of admixture between North-European and Inuit populations that deserves to be further explored.
The present work focused on the analysis of autosomal and X-chromosomal data in Greenland, aiming to increase the knowledge of the history and diversity among Greenlanders. Owing to its mode of inheritance, the X chromosome is expected to retain signs of linkage disequilibrium (LD) for longer periods of time than autosomes. Unlike the analysis of mtDNA and the Y chromosome, which inform about the history of female or male lineages, respectively, the autosomes and the X chromosome allow the simultaneous study of both sexes. Moreover, the fact that males only have one copy of the X chromosome provides direct access to their haplotypes,12 which is an advantage for the direct study of LD when compared with autosomes. Taken together, the results show that genetic drift and a differentiated settlement history around Greenland were responsible for shaping the patterns of diversity observed in Greenlanders.
Materials and methods
Autosomal STR markers
Population samples
DNA of 867 unrelated individuals from Greenland (N=505) and Denmark (N=362) were analysed. The Greenlandic sample was comprised of individuals born in Greenland who self-identified as Greenlanders. This sample was further divided into 17 groups according to the place of birth of the individuals (see Figure 1). Subsamples with <10 individuals (from Qaanaaq, Kangaatsiaq and Illoqqortoormiut) were only used in the pooled Greenlandic sample.
Autosomal STR genotyping
DNA samples were analysed for 15 autosomal STR markers using the AmpFℓSTR Identifiler PCR Amplification kit (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s recommendations.
X-chromosomal markers
Population samples
A total of 477 male DNA samples from Greenland (N=312) and Denmark (N=165) were selected for this study. Moreover, X-chromosomal markers from two Asian populations – Bangladesh (N=58) and Taiwan (N=22) – were also studied.
Genotyping of X-chromosomal markers
The samples were genotyped for three different sets of X-chromosomal markers: (i) 12 X-STRs included in the Investigator Argus X-12 kit (Qiagen GmbH, Hilden, Germany); (ii) a set of 33 insertion–deletion polymorphisms (indels), 32 of which were previously reported by Pereira et al;13 and (iii) a set of 25 X-chromosomal SNPs.14 The number of individuals analyzed is presented in Table 1 (references for previously published data are also presented). Information on X-chromosomal marker names and their physical and genetic locations is provided in Supplementary Table S1.
Ethical approval
All samples involved in the study were anonymised DNA extracts previously obtained from healthy individuals from paternity cases from the Section of Forensic Genetics, Department of Forensic Medicine, Faculty of Health and Medical Sciences, University of Copenhagen, Denmark. For autosomal markers, the alleged parents were included in the study. In the case of X-chromosomal markers, the alleged fathers and the sons were selected. The current study was authorised and approved by the ethical committee of the Capital Region of Denmark (De Videnskabsetiske Komiteer for Region Hovedstaden: KF-01-037/03, H-1-2011-081 and H3-2012-023) and complies with the ethical principles of the 2000 Helsinki Declaration of the 206 World Medical Association (http://www.uma.net/e/policy/b3.htm).
Statistical analyses
Intrapopulation variability
Allele frequencies, mean numbers of pairwise differences and expected heterozygosities of autosomal and X-chromosomal markers were estimated using the Arlequin v.3.5 software.15 Deviations from Hardy–Weinberg expectations (HWE) for autosomal markers were also investigated with the Arlequin v.3.5 software15 using 1 000 000 Markov chain steps. No tests for HWE were performed on X-chromosomal markers, as only male individuals were analysed. Significant association between pairs of loci in X-chromosomal male haplotypes was tested with an exact test of LD,16 as implemented in the Arlequin v.3.5 software.15 The Markov chain analysis used 1 000 000 steps.
Genetic structure and comparison with other populations
Analysis of molecular variance (AMOVA) was performed taking into account the 14 Greenlandic subgroups. Pairwise FST values were calculated for the Greenlandic and Danish populations. Both analyses were carried out in Arlequin v.3.5 software15 using autosomal data. The significance of the fixation indices was tested with 10 000 permutations of the Markov chain. Allele frequencies of 13 autosomal STRs (CSF1PO, D13S317, D16S539, D18S51, D21S11, D3S1358, D5S818, D7S820, D8S1179, FGA, TH01, TPOX and vWA) in Greenland and Denmark were compared with the frequencies in other populations. Data for 26 worldwide populations were downloaded from the autosomal STR DNA database (http://www.strdna-db.org). Comparisons were also made using X-chromosomal data for X-STRs, X-SNPs and X-indels. More information on the populations used in the comparative analyses is provided in Supplementary Table S2. For both autosomal and X-chromosomal data, pairwise FST values were calculated from allele frequencies using the Poptree2 software.17 Multidimensional scaling (MDS) analyses based on the pairwise FST values were carried out in STATISTICA (Statsoft, Tulsa, OK, USA; http://www.statsoft.com). Whenever necessary, the correction for multiple tests was performed following the Holm–Šidák procedure.18
Isolation by distance
The correlation between genetic and geographic matrix distances was studied in the isolation-by-distance (IBD) software v.1.5.2,19 using pairwise FST from autosomal data and the geographic distances in kilometres (km). Two possible scenarios of dispersion were considered, one using the minimum surface distances and the other coastal distances between the sample locations. The significance of the correlation was tested using a Mantel test with 100 000 randomisations. Rousset’s genetic distances,20 calculated as FST/(1−FST), were plotted for both scenarios and a reduced major axis regression was used to calculate the regression parameters (slope, intercept and R2).
Population structure
The genetic structure of the Greenlandic population was further analysed using a Bayesian method implemented in the software STRUCTURE v.2.3.4.21 Analyses were carried out for both autosomal and X-chromosomal markers using 1 000 000 steps of burn-in followed by 1 000 000 repetitions for the MCMC; the ‘admixture’ and ‘correlated allele frequencies’ models were considered. The most probable number of populations was given by the posterior probability values, ln Pr(X|K). The number of clusters (K) considered in the analyses ranged from 1 to 5 and 1 to 3 (for autosomal and X-chromosomal markers, respectively). Only results for K=2 are presented; results for K>2 can be found in Supplementary material. For each K value, three independent runs were performed. The Danish population was also included to provide a reference for the European ancestry. For X-chromosomal markers, the results from two Asian populations (Bangladesh and Taiwan) were also included. Cluster membership coefficients (Q) were visualised using the software programmes CLUMPP v.1.1.222 and Distruct v.1.1.23
Results and discussion
Genetic diversity and HWE
Allele frequency data for the autosomal and X-chromosomal markers studied in the Danish, Greenlandic and two Asian populations are shown in Supplementary Tables S3–S6. No significant departure from HWE was detected for the majority of autosomal STRs, with the exception of D19S433 in Danes and D5S818 in the Greenlandic population in Aasiaat (after Holm–Šidák correction). Values of average gene diversities over loci and mean numbers of pairwise differences (and their standard deviations) are presented in Table 1. The lowest values of intrapopulation diversity for both autosomal and X-chromosomal markers were consistently found in the Greenland population (Table 1). The populations of Ammassalik and Nanortalik showed the lowest values of heterozygosity for autosomal STRs (0.654 and 0.668, respectively), whereas Narsaq (0.737) and Paamiut (0.742) were among the most diverse populations in Greenland (Supplementary Table S3). The reduced diversities found in Greenland were in agreement with previous studies.4, 24 This result can be explained by the past and present demography of the Greenlanders, in which small populations are isolated owing to the remoteness of the geographical location they inhabit and the limited transportation links between locations.
LD analysis
Analysis of pairwise LD for 70 X-chromosomal markers in male individuals from Denmark, Greenland, Bangladesh and Taiwan revealed a higher level of LD in the population of Greenland than in the Danish and Asian populations (in accordance with previous publications24). Seventeen out of 2415 pairs of loci showed statistically significant allelic association in Greenland (after Holm–Šidák correction). In contrast, Denmark and Bangladesh showed significant levels of LD in three pairs of loci and no markers were found to be in LD in the Taiwanese population. The results presented here are consistent with previous studies based on uniparentally inherited markers and with the complex history of Greenlanders. As mentioned previously, owing to its mode of inheritance, the X-chromosome retains higher levels of LD for longer periods of time.12 Moreover, owing to the different number of X chromosomes present in each sex, the X chromosome will tend to reflect primarily the population history of females. Studies based on mtDNA have highlighted the absence of European lineages in Greenland and the possible existence of bottlenecks through its history.4, 10 The reduction of effective population size, at least in females, and the population differentiation among Greenlandic sub-populations could partially explain the levels of LD observed in this study, although it is difficult to evaluate the relative contribution of each event.
Genetic structure of the Greenlandic population and comparison with other populations
To investigate possible signs of genetic structure in Greenland, an AMOVA analysis was performed with 14 Greenlandic sub-populations using autosomal data. The results showed that the majority of variation was detected within populations (98.9%) and a significant FST value was observed (FST=0.011; P<0.0001). Pairwise FST genetic distances were calculated between the 14 Greenlandic and Danish populations (Supplementary Table S7). Within Greenland, the populations in Ammassalik and Nanortalik presented the greatest number of significant genetic distances compared with the other populations, although they did not differ significantly from each other. A second AMOVA analysis was carried out excluding the sample from Ammassalik. The global FST decreased to around half of that of the previous value (FST=0.005) and was statistically nonsignificant (P>0.05).
The highest genetic distance was observed between the population of Denmark and Ammassalik (FST=0.101, P<0.0001). It is interesting to note that the degree of differentiation of 10% that was observed between the two populations is in the range of FST values that are commonly reported between major human population groups for autosomal markers.25 The Danish population also differed significantly from all other Greenlandic sub-populations, with pairwise FST values varying from 4 to 9% (P<0.0001). The FST value obtained among the Danes and the pooled Greenlandic population was 0.055 (P<0.0001). This indicates the striking genetic differences between the Greenlandic population, particularly Ammassalik, and the Europeans, represented here by the Danish population.
Pairwise FST genetic distances were also calculated for the sets of X-chromosomal markers analysed in the populations from Denmark, Greenland, Bangladesh and Taiwan (Supplementary Table S8). Although all FST values were statistically significant (P<0.0001), the highest genetic distances were found between Danes and Greenlanders, with the exception of X-SNPs, where the greatest differentiation involved Greenland and the two Asian populations. Overall, the results are in line with those derived from autosomal data, revealing higher values of FST between Danes and the Greenlanders. Because of the haploid state in males, genetic drift proceeds more rapidly on the X chromosome, creating greater differentiation across populations on the X chromosome than on the autosomes.12 The available data only allowed the comparison of the pairwise genetic distances provided by STRs. Furthermore, several factors can complicate the comparison, including gene diversity of the loci used for both data sets and differences in maternal and paternal generation intervals and gene flow. The FST value observed was slightly higher for X-chromosomal STRs, although its order of magnitude was essentially the same as that derived from autosomal STRs (0.059 versus 0.055 for the X chromosome and autosomes, respectively).
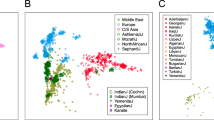
To contextualise the genetic diversity of the populations studied in a broader geographical area, the results were compared with previously reported data (Supplementary Table 2). Pairwise FST based on 13 autosomal STRs are represented in an MDS plot (Figure 2). The figure illustrates the clustering of European, African and Asian populations and the position of Danes within the framework of European diversity. The 14 Greenlandic populations were distant from most groups and appear to be more related to the Inuit from Alaska, especially the Inupiat and the Yupik. The genetic differentiation between these Alaskan samples and Athabaskan, also from Alaska, was previously addressed in Budowle et al26 and was attributed to the genetic and linguistic relationships between the Athabaskan and Native American groups like Navajos and Apaches. The sample of Native Americans included in the analysis presented an intermediate position between Europeans and Inuit, which is also an indication of their admixed origin. The relative locations in the plot of all Greenlandic populations reinforce previous results and historic accounts that the founder Inuit groups determine the genetic background of Greenlanders.
Multidimensional scaling analysis based on pairwise FST genetic distances calculated for 13 autosomal STRs. Population codes are as follows: AAS, Aasiaat; DK, Denmark; ALK_A, Alaska (Athabaskan); ALK_I, Alaska (Inupiat); ALK_Y, Alaska (Yupik); AMM, Ammassalik; CHI, China (Hong Kong); CHI_B, China (Beijing); GUI, Guinea Bissau; ILU, Ilulissat; JAP, Japan; MAN, Maniitsoq; MON_H, Mongolia (Hmar); MON_L, Mongolia (Lusei); MON_La, Mongolia (Lai); MON_M, Mongolia (Mara); MOR, Morocco; MOZ, Mozambique; NAMs, Native Americans; NAN, Nanortalik; NAR, Narsaq; NMB, Namibia; NUU, Nuuk; PAA, Paamiut; POL, Poland; POR, Portugal; QAQ, Qaqortoq; QAS, Qasigiannguit; QEQ, Qeqertarsuaq; RUS_K, Russia (Kemerovo); RUS_S, Russia (Saratov); SAR, Saudi Arabia; SIS, Sisimiut; SKOR, South Korea; SOM, Somalia; SPA, Spain; SWE, Sweden; TAI, Taiwan; TAN, Tanzania; UPE, Upernavik; UUM, Uummannaq.
For the X-chromosomal data, an MDS analysis was separately conducted for each set of X markers (Supplementary Figure S1). Once again, data clustered according to the continent of origin. Independently of the number or type of markers considered, the Danes were positioned among the set of European samples and the Greenlanders appeared systematically distant from all other populations.
Isolation by distance
To investigate whether the results obtained for the FST analyses were due to the isolation of the sub-populations in Greenland or could be explained by other factors (such as differences in founder populations), simulations were performed with the IBD software19 for the 14 Greenlandic populations using autosomal data. Two possible scenarios were considered: (i) dispersion along the coastline or (ii) dispersion across the inland ice cap. When negative distances are present, the software can set all negative values to zero (‘linear scale’) or use a logarithmic transformation of the values (‘logarithmic scale’). Owing to the similarity of the results obtained, only data for the linear scale model are presented (Table 2), but additional results can be found in Supplementary Figure S2.
IBD is the decrease in the genetic similarity among populations as the geographic distance between them increases.27 Indeed, the positive slope values obtained for the two dispersion scenarios conformed to that expectation of the presence of IBD. Nevertheless, significant P-values (P<0.05) were only obtained in the Mantel test for the correlation with coastal geographic distances. This result is understandable given the difficulty of transportation across the island, as the surface is mostly covered with ice. As a consequence, migration of individuals and communication between the various communities occur primarily along the coastline, mainly on the Western coast where most of the Greenlanders reside. According to the reduced major axis regression correlation that is implemented in the IBD software,19 approximately 84% of the genetic differentiation inside the Greenlandic sub-populations may be due to a combination of effects, most likely genetic drift, and/or a different degree of non-Inuit gene flow within the different subgroups (Table 2). The remaining 15.7% of differentiation could be ascribed to geographical distances. Nevertheless, because the sub-population from Ammassalik presented the lowest levels of genetic diversity, another test was carried out excluding this subsample as an attempt to assess how this population contributed to the observed results. When the individuals from Ammassalik were not considered, the role of IBD in creating genetic differentiation among groups was approximately 2.8%, revealing that factors other than geographic distance might be responsible for the genetic differentiation of the Greenlandic sub-populations.
Population admixture
To address the extent and relative contribution of Northern European ancestry in the population of Greenland, autosomal and X-chromosomal data were analysed in STRUCTURE v.2.3.4.21 From the estimated posterior probability of the data (ln Pr(X|K)) obtained after 15 independent runs from K=1 to 5, it was consistently inferred that the most probable number of clusters (K) was K=2. The membership coefficient of each individual to belong to each of the two clusters (Q values) is depicted in Figure 3. The results for K>2 are presented as Supplementary Information (Supplementary Figure S3). The mean and standard deviation of the proportion of membership of each sub-population to each cluster is also presented as Supplementary Information (Supplementary Table S9). Figure 3a clearly illustrates that most of the individuals from Ammassalik belonged to cluster 2 (light yellow), as did the majority of individuals from Nanortalik. Regarding the remaining Greenlandic sub-populations from South and West Greenland, the results showed different levels of admixed ancestry between clusters 1 and 2. Cluster 2 is most likely representative of non-Danish ancestry, because most Danish individuals were assigned to cluster 1 (blue). These findings are in agreement with those obtained from the AMOVA and the pairwise FST values that also reflected the genetic differences between Greenlanders and Danes. A similar pattern of substructure in Greenland has previously been described based on the study of mtDNA.4 An analysis including only the 14 Greenlandic sub-populations (Supplementary Figure S3a) revealed different cluster proportions compared with the previous result. This is most likely due to the presence or absence of a reference European population in the analysis. Future studies should include other Inuit reference populations to clarify the differences observed when K>2.
Representation of the membership coefficients obtained from the STRUCTURE analysis. (a) Greenlandic and Danish populations studied at 15 autosomal STRs (K=2); (b) Greenlanders and Danes analysed with 70 X-chromosomal markers (K=2 and 3, also including Asians from Bangladesh and Taiwan). Admixture and correlated allele frequency models were taken into account for autosomal and X-chromosomal markers.
Analysis of the X-chromosome data (Figure 3b) was performed for Greenlanders and Danes (K=2) and the two Asian populations analysed in this work (K=3). The two Asian populations were assumed to be the best proxies for Inuit (as X-chromosomal data for other Inuit populations are currently unavailable), under the rationale that the comparison between the Greenlanders and the Asian groups could help to discern differences in the Northern European and Asian genetic heritages found among Greenlanders. The results mirrored those obtained for the autosomal STRs, with Greenlanders presenting a low level of the European/Danish ancestry component and also a low level of Asian ancestry. Taking into account that data for X-chromosomal markers includes individuals from all around Greenland, the proportion of European ancestry observed in X-chromosome markers (13%) is lower than that observed for autosomal data (27%) (Supplementary Table S9); the t-test P-value was statistically significant (P<0.0001). This result is not completely unexpected, because previous studies of uniparentally inherited markers4, 5, 6, 7, 10 reported that the matrilineal ancestry of the Greenlanders is mostly non-European.
Conclusions
Although previous reports exist on Greenlandic genetic diversity, this is the first study performing an analysis based on autosomal and X-chromosomal information. This work attested to the complexity of the genetic pool of Greenlanders, corroborating a number of previous reports.1, 4, 5, 6, 7, 8, 9, 10, 24 Taken as a whole, the results supported the historical and archaeological accounts of Greenland’s demographic features. A hard-to-access geography, together with an adverse climate, rendered difficult not only the most ancient peopling of Greenland, initiated a few millennia ago, but also the settlement of the island. The relative isolation restricted contact from other regions, and the immigration history paved by successive waves of migration could have created conditions for strong bottlenecks and other effects of genetic drift in the Greenlandic population. Signs of the intricate history are still visibly retained in the genetic patterns of current Greenlanders, as exemplified by the reduced levels of diversity and the increased extent of LD in comparison with other populations. Importantly, this work brought attention to the population of Ammassalik, which consistently exhibited the lowest levels of diversity and European admixture. The pattern of substructure detected in Greenland corroborates previous reports on mtDNA4, 10 and Y-chromosomal lineages6, 7, 8, 9 that indicated that East Greenland is the most isolated area in Greenland. It also suggests that Ammassalik most likely represents one of the best windows into the genetics of Greenlanders in pre-European times.
References
Gilbert MT, Kivisild T, Gronnow B et al: Paleo-Eskimo mtDNA genome reveals matrilineal discontinuity in Greenland. Science 2008; 320: 1787–1789.
McGhee R : Radiocarbon dating and the timing of the Thule migration; in Applet M, Berglund J, Gullov HC, (eds): Identities and Cultural Contacts in the Arctic. Copenhagen, Denmark: Danish National Museum & Danish Polar Center, 2000, pp 181–191.
Morrison D : The arrival of the Inuit: Amundsen Gulf and the Thule migration; in Applet M, Berglund J, Gullov HC, (eds): Identities and Cultural Contacts in the Arctic. Copenhagen, Denmark: Danish National Museum & Danish Polar Center, 2000, pp 221–228.
Helgason A, Palsson G, Pedersen HS et al: mtDNA variation in Inuit populations of Greenland and Canada: migration history and population structure. Am J Phys Anthropol 2006; 130: 123–134.
Kleivan I : History of Norse Greenland; in Damas D, (ed): Handbook of North American Indians, Vol 5. The Arctic. Washington DC, USA: Smithsonian Institution, 1984; 549–555.
Bosch E, Calafell F, Rosser ZH et al: High level of male-biased Scandinavian admixture in Greenlandic Inuit shown by Y-chromosomal analysis. Hum Genet 2003; 112: 353–363.
Bosch E, Rosser ZH, Norby S et al: Y-chromosomal STR haplotypes in Inuit and Danish population samples. Forensic Sci Int 2003; 132: 228–232.
Sanchez JJ, Børsting C, Hernandez A et al: Y chromosome SNP haplogroups in Danes, Greenlanders and Somalis. Int Congr Ser 2004; 1261: 347–349.
Hallenberg C, Tomas C, Simonsen B et al: Y-chromosome STR haplotypes in males from Greenland. Forensic Sci Int Genet 2009; 3: e145–e146.
Saillard J, Forster P, Lynnerup N et al: MtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet 2000; 67: 718–726.
Rasmussen M, Li Y, Lindgreen S et al: Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature 2010; 463: 757–762.
Schaffner SF : The X chromosome in population genetics. Nat Rev Genet 2004; 5: 43–51.
Pereira R, Pereira V, Gomes I et al: A method for the analysis of 32 X chromosome insertion deletion polymorphisms in a single PCR. Int J Legal Med 2012; 126: 97–105.
Tomas C, Sanchez JJ, Barbaro A et al: X-chromosome SNP analyses in 11 human Mediterranean populations show a high overall genetic homogeneity except in North-west Africans (Moroccans). BMC Evol Biol 2008; 8: 75.
Excoffier L, Lischer HE : Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 2010; 10: 564–567.
Raymond M, Rousset F : An exact test for population differentiation. Evolution 1995; 49: 1280–1283.
Takezaki N, Nei M, Tamura K : POPTREE2: Software for constructing population trees from allele frequency data and computing other population statistics with Windows interface. Mol Biol Evol 2010; 27: 747–752.
Holm S : A simple sequentially rejective multiple test procedure. Scand J Stat 1979; 6: 65–70.
Bohonak AJ : IBD (isolation by distance): a program for analyses of isolation by distance. J Hered 2002; 93: 153–154.
Rousset F : Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 1997; 145: 1219–1228.
Pritchard JK, Stephens M, Donnelly P : Inference of population structure using multilocus genotype data. Genetics 2000; 155: 945–959.
Jakobsson M, Rosenberg NA : CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007; 23: 1801–1806.
Rosenberg NA : Distruct: a program for the graphical display of population structure. Mol Ecol Notes 2004; 4: 137–138.
Tomas C, Pereira V, Morling N : Analysis of 12 X-STRs in Greenlanders, Danes and Somalis using Argus X-12. Int J Legal Med 2012; 126: 121–128.
Jobling MA, Hurles M, Tyler-Smith C : Human Evolutionary Genetics: Origins, Peoples and Disease Kentucky, USA 2003.
Budowle B, Chidambaram A, Strickland L et al: Population studies on three Native Alaska population groups using STR loci. Forensic Sci Int 2002; 129: 51–57.
Jensen JL, Bohonak AJ, Kelley ST : Isolation by distance, web service. BMC Genet 2005; 6: 13.
Pereira V, Tomas C, Sanchez JJ et al: Study of 25 X-chromosome single nucleotide polymorphisms in African and Asian populations. Forensic Sci Int Gen Suppl Ser 2011; 3: e139–e140.
Acknowledgements
We thank all the personnel at the Department of Forensic Medicine in Copenhagen who were involved in the sample collection and typing. We also thank the Unidade de Xenética, Instituto de Ciencias Forenses and Departamento de Anatomía Patolóxica e Ciencias Forenses, Universidade de Santiago de Compostela for providing samples from Taiwan. This work was partially supported by Ellen and Aage Andersen’s Foundation. IPATIMUP is an Associate Laboratory of the Portuguese Ministry of Science, Technology and Higher Education and is partially supported by FCT.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Pereira, V., Tomas, C., Sanchez, J. et al. The peopling of Greenland: further insights from the analysis of genetic diversity using autosomal and X-chromosomal markers. Eur J Hum Genet 23, 245–251 (2015). https://doi.org/10.1038/ejhg.2014.90
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2014.90
This article is cited by
-
Genetic characterization of 32 X-InDels in a population sample from São Paulo State (Brazil)
International Journal of Legal Medicine (2019)
-
A racial classification for medical genetics
Philosophical Studies (2018)
-
Determination of NAT2 acetylation status in the Greenlandic population
Archives of Toxicology (2016)