Abstract
MRXS5 or Pettigrew syndrome was described 20 years ago in a four generation family including nine affected individuals presenting with facial dysmorphism, intellectual disability, Dandy–Walker malformation and inconstant choreoathetosis. Four individuals had iron deposition in the basal ganglia seen on MRI or at autopsy. The mutation causing Pettigrew has remained elusive since the initial description of the condition. We report the identification of a mutation in the X-linked AP1S2 gene in the original Pettigrew syndrome family using X-chromosome exome sequencing. We report additional phenotype details for several of the affected individuals, allowing us to further refine the phenotype corresponding to this X-linked intellectual disability syndrome. The AP1S2 c.426+1 G>T mutation segregates with the disease in the Pettigrew syndrome family and results in loss of 46 amino acids in the clathrin adaptor complex small chain domain that spans most of the AP1S2 protein sequence. The mutation reported here in AP1S2 is the first mutation that is not predicted to cause a premature termination of the coding sequence or absence of the AP1S2 protein. Although most of the families affected by a mutation in AP1S2 were initially described as having different disorders assigned to at least three different OMIM numbers (MIM 300629, 300630 and 304340), our analysis of the phenotype shows that they are all the same syndrome with recognition complicated by highly variable expressivity that is seen within as well as between families and is probably not explained by differences in mutation severity.
Similar content being viewed by others
Introduction
X-linked intellectual disability (XLID) accounts for up 10% of males with intellectual disability.1 A large number of genes whose mutations cause syndromic or nonspecific XLID have been identified during the last two decades,2 demonstrating a high degree of genetic and phenotypic heterogeneity. However, the most recent review suggested that the genetic cause of 79 syndromic forms of XLID (from an estimated 160) have not yet been identified.1 X-linked Dandy–Walker malformation with mental retardation (one would now write ‘intellectual disability’), basal ganglia disease and seizures (MIM 304340), also designated as MRXS5 or Pettigrew syndrome, was described 20 years ago in a four generation family including nine affected individuals.3 Patients had facial dysmorphism with a long and narrow face, intellectual disability, Dandy–Walker malformation (but see results below) and inconstant choreoathetosis. Four individuals had iron deposition in the basal ganglia seen on MRI or at autopsy. Head circumference was highly variable with both microcephaly and macrocephaly being reported in affected individuals from the original family.
Pettigrew syndrome was mapped to Xq26 with a lod score of 2.19,4 although a recurrence subsequent to this report showed that the mapping data were possibly not correct. Despite several attempts to identify the causative gene, the mutation causing Pettigrew syndrome has remained elusive since the initial description of the condition. Here, we report the identification of a mutation in the AP1S2 gene—the same gene mutated in X-linked Fried syndrome—in the original Pettigrew syndrome family. We also report additional phenotype details for several of the affected individuals, allowing us to further refine the phenotype corresponding to this XLID syndrome.
Materials and methods
Ethical statement
A prior study of this family4 was approved by the Institutional Review Board at Baylor College of Medicine. Here we studied one affected individual (V.1) and his parents, who provided updated medical history on several relatives first reported in prior publications. Informed consent was obtained from the new patient and both parents. The study was approved by the Institutional Review Board at Seattle Children’s Hospital (IRB #13 291). Lymphoblastoid cell lines from individuals III.2, III.14, IV.4 and III.3b were obtained from the Coriell Cell Line Repository (Camden, NJ, USA; catalog numbers: GM12523, GM12534, GM12542 and GM12543).
Exome sequencing and reference sequence
The exonic sequences were captured with the Agilent Sure Select Human Exome V3 kit (Agilent, Santa Clara, CA, USA) and sequencing was performed on an Illumina HiSeq2000 sequencing apparatus (Illumina, San Diego, CA, USA) used with pipeline version 1.5 at Centre National de Génotypage (Evry, France). Sequences were mapped on human genome build hg19 with the commercial tool CLC Genomics Workbench version 4.9 (CLCBIO, Aarhus, Denmark). The SNP detection was also performed with CLCBIO. We kept variants with a minimum quality of 30 and a minimum average quality of surrounding bases of 20. The AP1S2 mutation is numbered according to reference sequence NM_003916.3. The AP1S2 mutation was submitted to the corresponding LOVD database at http://databases.lovd.nl/shared/variants/0000017199.
RNA extraction and reverse transcription
Total RNA was extracted from the lymphoblastoid cell lines of two affected males (III.2 and III.14), one unaffected male (IV.4) and one carrier mother (III.3b) using PerfectPure RNA Tissue kit (5 PRIME GmbH, Hamburg, Germany) according to the instructions of the manufacturer. The RNA preparations were analyzed for purity using the ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA). Reverse transcription of 2 μg of total RNA was performed in 20 μl of Superscript II reaction buffer (Invitrogen Corp., Carlsbad, CA, USA) containing 0.2 M dithiothreitol, 250 ng of dN6, 40 U of Rnase inhibitor (Invitrogen Corp.), 10 mM dNTP and 200 U of Superscript II reverse transcriptase (Invitrogen Corp.).
Classical PCR and sequencing
To amplify the region containing the c.426+1 G>T mutation (exon 4) on genomic DNA, we performed PCR reactions in a total volume of 50 μl, containing 1 × PCR buffer, 0.2 mM dNTPs, 1.5 mM MgCl2, 1 U Taq polymerase, 200 nM of each primer and 50 ng of genomic DNA. The primers used were located in intron 3 (forward, 5′-AGAGCCCAGTCTTATTTAGC-3′) and intron 5 (reverse, 5′-ATGGACACTACATGACAAGC-3′) of AP1S2 producing a 480-bp PCR product. The program included an initial denaturation (94 °C, 3 min) followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s.
To amplify the transcript revealing the skipping of AP1S2 exon 4 in the patient’s cDNAs, we performed PCR reactions in a total volume of 50 μl, containing 1 × PCR buffer, 0.2 mM dNTPs, 1,5 mM MgCl2, 1 U Taq polymerase, 200 nM of each primer and 1 μl of cDNA produced using the patient’s RNA (see above). Primers used were located in exon 3 of the gene (forward, 5′-CTGTGCTATTGAGGATCAGG-3′) and exon 5 (reverse, 5′-CATCATCTTACACCATGAGC-3′) producing a 368-bp PCR product in a control cDNA and a 230-bp PCR product in the patient cDNA (Figure 3b, left panel); or located in exon 4 of the gene (forward, 5′-CTTAAAGCAATTGAGCAGGC-3′) and exon 5 (reverse, 5′-CATCATCTTACACCATGAGC-3′) producing a 178-bp PCR product in control cDNA and no amplification in patient cDNA (Figure 3b, right panel). The program included an initial denaturation (94 °C, 3 min) followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s.
Results
Clinical findings
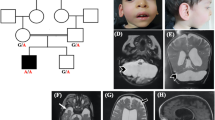
The pedigree of the original Pettigrew syndrome family is presented in Figure 1, modified only slightly from the original published pedigree.
Pedigree of the studied family. For clarity purpose, the numbering of the individuals in the present pedigree were kept identical to the initial description of Pettigrew et al,3 with individuals 2b, 3b and 7b being individuals 2, 3 and 7 in Pettrigew et al.3 An affected male born in the interval was added in generation V. Individuals for whom a DNA sample is available are represented with a dash above their symbol. Hatched symbols represent clinically affected individuals for whom a DNA sample was not available to perform molecular screening. Obligate carrier females are indicated.
Patient III.8
This man had early developmental delay, poor feeding and lethargy or reduced activity (‘listless’) in infancy. He walked at 26 months, learned to use a few words in childhood that he later lost, and had no self help skills. He developed self-abusive behaviors, mostly slapping himself. Examination showed mildly coarse facial features (see Figure 2d in Pettigrew et al3). He became increasingly spastic, developed slow athetoid movements, severe scoliosis and proximal contractures and was bedridden by his early 20s. At that age, his height was 175 cm (40th percentile) and OFC 50.1 cm (about –4 standard deviations). He had the onset of seizures at about 31 years and died, probably in his mid- to late 30’s. At autopsy, the frontal lobes were unusually small with a decreased neuron number. Excess iron was present in glial cells of the putamen and globus pallidus of the basal ganglia and in the substantia nigra, red nuclei and dentate nuclei. Axonal dystrophy was noted in the cerebellum and cerebrum. Iron was noted in the atrophic skeletal muscles, and hemosiderosis of the liver and spleen was present.
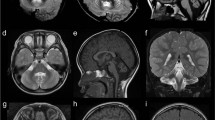
(A) Photograph of affected individual V.1 at 16 years of age shows a long face, broad forehead, deep-set eyes, large mouth with full lips and prominent jaw. (B) Midline sagittal T1-weighted MRI scans of patient V.1 at ages 1 (a), 3 (b) and 5 (c) years compared with a normal control (d). The first scan (a) shows a very large, fluid-filled posterior fossa and flattening of the posterior vermis (white arrow). The inferior portion of the posterior vermis is abnormally small in all three scans (asterisks in a–c). (C) Serial brain imaging studies in patient V.1 including axial MRI T2-weighted MRI images at 1 (a), 3 (b) and 5 (c) years, and in a normal control (d) for comparison; CT scans at 3 (e) and 10 (f) years of age; and inverted T2-weighted images at 5 years (g and h). The first scan (a) shows moderately enlarged 3rd and lateral ventricles and a large posterior fossa cyst (c). All CT and MRI images show normal basal ganglia (asterisks placed over the left putamen in a–c and e, f). Multiple periventricular nodular heterotopia are seen along the right frontal horn (arrow in b and g) and left mid-body (h) of the lateral ventricles.
Patient III.14
This man had developmental delay, hypotonia, poor head control and truncal ataxia in infancy. He walked at 2 years and used his first words at 18 months. Examination showed a prominent forehead and long face (see Figures 2e and f in Pettigrew et al3). At 39 years, he was able to walk, feed and dress himself with help, and understand simple commands, repeat short phrases with echolalia and perseveration. He had spasticity, kyphoscoliosis with stooped posture and mild proximal contractures. His weight was 55 kg (3–10th percentile), height 175 cm (25–50th percentile) and OFC 56.6 cm (75th percentile).
Patient IV.8
This man was born at term and had a small anterior fontanelle. Developmental delay and hypotonia were apparent from the first few months of life. At 20 months, he functioned at a 6-month developmental level. By 3 years, he was able to sit with support and crawl. Scissoring of the lower limbs and brisk deep tendon reflexes were present, although overall tone was decreased. General athetoid movements were noted at 4 years, and self-abuse in the form of face slapping began at 5–6 years. By 7 years, he walked independently with a broad-based, unsteady gait. An EEG at 10 years showed slow background, and the Vineland Social Maturity Scale demonstrated profound intellectual disability with a functional age of 13 months. On examination at 25 years, his height was 178 cm (60th percentile), weight 48 kg (∼5th percentile) and OFC was 55 cm (25th centile). Pertinent findings included dolichocephaly, long narrow face, coarse facial appearance with prominent supraorbital ridges, right esotropia, large nose with a broad bulbous tip, large mouth with thick patulous lips and prominent mandible (see Figure 2a in Pettigrew et al3). Neurological abnormalities included spasticity of upper and lower limbs; mild intention tremor of the hands; wide-based, slow, shuffling gait; and brisk deep tendon reflexes. He fell frequently and lacked protective reflexes. He slapped his face often and was nonverbal. An MRI scan of the brain showed mild dilation of the fourth ventricle but no hydrocephalus or cerebellar hypoplasia, as well as abnormal signal consistent with increased iron deposition in the substantia nigra, globus pallidus, dentate nuclei and red nuclei. At 46 years of age, he has difficulty walking and loses abilities with illnesses. He never talked and never went to school.
Patient V.1
This boy was born several years after the original report. Neonatal records were normal, but his early development was delayed. He sat and began to use a few words just before 12 months, pulled to sit by 2 years and walked by 3 years. Language development was delayed. Hydrocephalus detected after brain MRI at 15 months was shunted. His growth was slow the year after his shunts were placed, but thereafter his height has remained between the 25th and 50th percentiles, and his weight between the 50th and 75th percentiles. His parents reported frequent hand flapping behaviors in childhood. These mostly resolved by 12 years of age, but he still rocks side-to-side at times, and when excited has some extensor posturing. When examined at 16 years (Figure 2a), his head circumference was 57.0 cm (75th percentile). He had mild ptosis possibly related to fatigue and full lips. His neurological exam revealed limited social interaction, poor eye contact and generally reduced movements. He was able to speak in single words and short sentences. His language comprehension was better than his spoken language, and he was able to follow simple directions including some two-step directions. His stance was mildly unsteady, and his gait was slow with his arms held together on his abdomen while walking. Serial brain imaging studies showed cerebellar vermis hypoplasia (CBVH), an initial very large posterior fossa and mega-cisterna magna that became much smaller after his initial shunt, moderately enlarged third and lateral ventricles, and a cluster of periventricular nodular heterotopia on the left (Figure 2). No calcifications or iron were seen, with his last MRI done at 4.5 years and his last CT scan at 10 years of age.
Mutation identification
Next generation sequencing technologies have proven useful to identify mutations in a wide range of genetic diseases, including intellectual disability,5 in cases where classical candidate gene approaches (whether functional or positional) were not successful. We analyzed the human X-chromosome exome in one affected individual (IV.8) from the original Pettigrew syndrome family (Figure 1). A summary of the sequencing results is presented in Supplementary Table 1. All SNPs were filtered against dbSNP132 and we discarded variations for which we had less than 10 reads or that were located outside coding sequences or farther than ±5 nucleotides from an exon. Candidate SNPs were then analyzed using the Universal Mutation Database High Throughput Sequencing interface6 at http://www.umd-hts.eu to prioritize potentially disease causing SNPs. Candidate variants were confirmed using Sanger sequencing and their segregation was analyzed in the family. This process led to the identification of a c.426+1G>T mutation in exon 4 of the gene encoding the sigma 2 subunit of the adaptor protein 1 complex (AP1S2) in Xp22 (Figure 3a). The mutation segregates with the disease in all tested individuals (III.2, III.3b, III.5b, III.14, IV.7b, IV.8 and V.1, Figure 1). The AP1S2 gene is located in Xp22, a region of the human X chromosome that had been excluded in the previously published linkage study.4 Nonetheless, we found that the c.426+1 G>T mutation does segregate with the disease, suggesting that exclusion of the Xp21-p22 region in the initial linkage analysis was incorrect.
Characterization of the mutation and its consequence for the AP1S2 transcript and protein. (a) Sanger sequencing of the region containing the c.426+1 G>T mutation (arrow) in an affected male (IV.8), his carrier mother (III.5b) and an unaffected male from the same family (IV.4). The sequence belonging to exon 4 of AP1S2 is labelled. (b) RT-PCR amplification of the AP1S2 transcript using primers located in exons 3 to 5 (left) or 4 and 5 (right) in samples prepared from the lymphoblastoid cell lines of affected individuals (III.2 and III.14), a carrier female (III.3b) and an unaffected male (IV.4). A control sample prepared from the lymphoblastoid cell line of a normal individual was included (C+) together with a negative control (no DNA; C−). The size of the PCR amplicons is indicated. The molecular weight marker is the 1 kb+ DNA ladder (Invitrogen). (c) Schematic representation of the consequence of the c.426+1 G>T mutation, putatively leading to the deletion of 46 amino acids from the AP1S2 protein (p.Val97_Glu142del; indicated by the minus signs below the protein sequence).
The c.426+1G>T mutation is predicted to disrupt normal splicing of the AP1S2 gene. In order to determine the consequence of the c.426+1G>T mutation for the AP1S2 transcript, we performed an RT-PCR experiment using cDNA prepared from lymphoblastoid cell lines from two affected individuals (III.2 and III.14), an obligate carrier female (III.3b) and an unaffected male (IV.4). This experiment revealed a transcript of abnormal size in the patients’ cDNAs (Figure 3b). Sequencing of the abnormal PCR product showed that the c.426+1G>T mutation causes the skipping of AP1S2 exon 4 and the junction of exons 3 and 5 in the mutant cDNA (mutation nomenclature at the RNA level is r.288_426del according to sequence NM_003916.3). The patients (III.2 and III.14) have a single cDNA species of abnormal size, whereas a carrier female (III.3b) produces a vast majority of wild-type transcript, although a very faint band of abnormal size can be seen if the gel is overexposed. X-chromosome inactivation studies demonstrated a highly skewed pattern at 95:5 for female III.3b (data not shown). Other female carriers in this family have either a random pattern of XCI (III.5b) or could not be tested (female IV.7b is homozygous at the loci used to assess XCI). Skewing of X-chromosome inactivation in female III.3b could be due to clonal expansion during the expansion of the lymphoblastoid cell line given that the majority of females with a heterozygous AP1S2 mutation have a random XCI pattern. The junction of exons 3 and 5 in the AP1S2 transcript maintains the open reading frame but results in the deletion of 46 amino acids from the AP1S2 protein sequence (p.Val97_Glu142del; Figure 3c). Hence, if a mutant protein is synthesized, it would have a reduced size of 111 amino acids compared with the 157 amino-acid wild-type protein.
Discussion
AP1S2 encodes the sigma-2 subunit of the heterotetrameric adaptor protein 1 (AP1) complex. AP1 is found in the cytosolic side of coated vesicle in the Golgi compartment. It mediates the recruitment of clathrin and the recognition of sorting signals of transmembrane receptors.7 XLID associated with mutations in AP1S2 (MIM 300629) was first described in 2006 in three families with mild to profound XLID, including two families previously reported with nonspecific XLID or syndromic XLID with marked hypotonia in infancy, severe intellectual disability and later ‘difficult’ behavior.8, 9, 10 The phenotype was expanded in a report of two families including the original Scottish family with Fried syndrome (MIM 300630) with highly variable XLID as well as hydrocephalus and basal ganglia calcifications.11, 12, 13 Elevated CSF protein levels were also added to the clinical description,14 although only two patients were analyzed for that specific biochemical characteristic.
The disorder studied here was first reported in 1991 as XLID with Dandy–Walker malformation, basal ganglia disease and seizures,3 and subsequently designated as MRXS5 or Pettigrew syndrome (MIM 304340). As initially described, this disorder differs from the AP1S2-associated disorder designated XLID, Fried type (MIM 300630) because of several key features including Dandy–Walker malformation, hydrocephalus, choreoathetosis and iron deposition in the basal ganglia.
Although most of these families were initially described as having different disorders assigned to at least three different OMIM numbers (OMIM 300629, 300630 and 304340), our analysis of the phenotype shows that they are all the same syndrome with recognition complicated by highly variable expressivity that is seen within as well as between families and is likely not explained by differences in mutation severity. We identified more than 20 features of this syndrome across the seven reports supplemented by our evaluation of patient V-I (this report), and these are listed in Supplementary Tables 2 and 3. The abnormalities include a variable but often severe developmental disorder from infancy that is complicated by later onset of neurodegeneration.
Brain imaging reveals variable anomalies that may include normal brain imaging at least in early childhood. The developmental abnormalities may include microcephaly that is probably postnatal, hydrocephalus11, 12 (and this report), periventricular nodular heterotopia (novel in this report) and cerebellar–posterior fossa anomalies. The cerebellar and posterior fossa defects were barely evaluated in prior reports, but at least one individual had CBVH13 and another had a ‘posterior arachnoid cyst’ seen on CT scan10 that we suspect was not a normal variant. In the family reported here, we found evidence for cerebellar malformations across the Dandy–Walker malformation spectrum that include CBVH with severe mega-cisterna magna (patient V.I) and probably classic Dandy–Walker malformation based on several brain imaging reports. The Dandy–Walker malformation could be linked to the hydrocephalus reported in many affected individuals.
Reports of calcium deposits in Fried syndrome and iron deposition in Pettigrew syndrome are confusing. No tissue is available for us to confirm the earlier report of iron deposition in the basal ganglia, which is important given the convincing images of calcium shown on several brain CT scans. However, both the iron and calcium depositions involve regions of the brain where iron levels are the highest (basal ganglia and cerebellar dentate nuclei). The most parsimonious explanation is that the neurodegeneration begins with iron deposition, which was seen on both brain pathology and imaging in the Pettigrew syndrome family. The calcifications probably follow iron deposition, with onset in childhood most likely between 1 and 9 years (based on patient 2 in Borck et al study14). However, this proposed progression needs to be documented on serial brain imaging studies.
Interestingly, it appears that most female carriers of a mutation in the AP1S2 gene and tested for X-chromosome inactivation have a random pattern of XCI (with the exception of female III.3b reported here) and are described as having normal intelligence,9 even when the whole AP1S2 gene is deleted.15 Although X-chromosome inactivation has not been tested in the brain, these observations suggest that a proportion of normal cells in a mosaic context is sufficient to prevent carrier females from being intellectually disabled in the presence of an AP1S2 mutation.
The finding that Pettigrew syndrome is caused by AP1S2 mutation further contributes to the description of the clinical features associated with mutations in this gene and provides a molecular cause for an additional XLID syndrome.
References
Lubs HA, Stevenson RE, Schwartz CE : Fragile X and X-linked intellectual disability: four decades of discovery. Am J Hum Genet 2012; 90: 579–590.
Tarpey PS, Smith R, Pleasance E et al: A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet 2009; 41: 535–543.
Pettigrew AL, Jackson LG, Ledbetter DH : New X-linked mental retardation disorder with Dandy-Walker malformation, basal ganglia disease, and seizures. Am J Med Genet 1991; 38: 200–207.
Huang TH-M, Hejtmancik JF, Edwards A et al: Linkage of the gene for an X-linked mental retardation disorder to a hypervariable (AGAT)-n repeat motif within the human hypoxanthine phosphoribosyltransferase (HPRT) locus (Xq26). Am J Hum Genet 1991; 49: 1312–1319.
Vissers LE, de Ligt J, Gilissen C et al: A de novo paradigm for mental retardation. Nat Genet 2010; 42: 1109–1112.
Frédéric MY, Lalande M, Boileau C et al: UMD-predictor, a new prediction tool for nucleotide substitution pathogenicity: application to four genes: FBN1, FBN2, TGFBR1, and TGFBR2. Hum Mutat 2009; 30: 952–959.
Kongsvik TL, Höning S, Bakke O, Rodionov DG : Mechanism of interaction between leucine-based sorting signals from the invariant chain and clathrin-associated adaptor protein complexes AP1 and AP2. J Biol Chem 2002; 277: 16484–16488.
Carpenter NJ, Brown WT, Qu Y, Keenan KL : Regional localization of a nonspecific X-linked mental retardation gene (MRX59) to Xp21.2-p22.2. Am J Med Genet 1999; 85: 266–270.
Tarpey PS, Stevens C, Teague J et al: Mutations in the gene encoding the Sigma 2 subunit of the adaptor protein 1 complex, AP1S2, cause X-linked mental retardation. Am J Hum Genet 2006; 79: 1119–1124.
Turner G, Gedeon A, Kerr B, Bennett R, Mulley J, Partington M : Syndromic form of X-linked mental retardation with marked hypotonia in early life, severe mental handicap, and difficult adult behavior maps to Xp22. Am J Med Genet A 2003; 117A: 245–250.
Fried K : X-linked mental retardation and/or hydrocephalus. Clin Genet 1972; 3: 258–263.
Strain L, Wright AF, Bonthron DT : Fried syndrome is a distinct X linked mental retardation syndrome mapping to Xp22. J Med Genet 1997; 34: 535–540.
Saillour Y, Zanni G, Des Portes V et al: Mutations in the AP1S2 gene encoding the sigma 2 subunit of the adaptor protein 1 complex are associated with syndromic X-linked mental retardation with hydrocephalus and calcifications in basal ganglia. J Med Genet 2007; 44: 739–744.
Borck G, Mollà-Herman A, Boddaert N et al: Clinical, cellular, and neuropathological consequences of AP1S2 mutations: further delineation of a recognizable X-linked mental retardation syndrome. Hum Mutat 2008; 29: 966–974.
Ballarati L, Cereda A, Caselli R et al: Deletion of the AP1S2 gene in a child with psychomotor delay and hypotonia. Eur J Med Genet 2012; 55: 124–127.
Acknowledgements
We wish to extend our sincere thanks to the original family with Pettigrew syndrome on both sides of the Atlantic, as they have borne with us for many years as we worked to solve this problem. We also thank Carlos Cardoso and Helen Kosakowski for their involvement in the initial phase of this work, Caroline Lacoste and Catherine Badens for X-chromosome inactivation analysis and the Centre de Ressources Biologiques of La Timone Children’s Hospital (Karine Bertaux, Cécile Mouradian and Andrée Robaglia-Schlupp) for assistance with the cell lines used in this study. This work was funded by the US National Institutes of Health under National Institute of Neurological Disorders and Stroke (NINDS) grant NS050375 (to WBD), and by Inserm and Aix Marseille University (to LV).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Cacciagli, P., Desvignes, JP., Girard, N. et al. AP1S2 is mutated in X-linked Dandy–Walker malformation with intellectual disability, basal ganglia disease and seizures (Pettigrew syndrome). Eur J Hum Genet 22, 363–368 (2014). https://doi.org/10.1038/ejhg.2013.135
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.135
Keywords
This article is cited by
-
MLKL deficiency protects against low-grade, sterile inflammation in aged mice
Cell Death & Differentiation (2023)
-
Golgipathies reveal the critical role of the sorting machinery in brain and skeletal development
Nature Communications (2022)
-
Genomics of human congenital hydrocephalus
Child's Nervous System (2021)
-
Intracranial calcifications in childhood: Part 2
Pediatric Radiology (2020)