Abstract
Hereditary retinal dystrophies (RD) constitute a group of blinding diseases that are characterized by clinical variability and pronounced genetic heterogeneity. The different forms of RD can be caused by mutations in >100 genes, including >1600 exons. Consequently, next generation sequencing (NGS) technologies are among the most promising approaches to identify mutations in RD. So far, NGS is not routinely used in gene diagnostics. We developed a diagnostic NGS pipeline to identify mutations in 170 genetically and clinically unselected RD patients. NGS was applied to 105 RD-associated genes. Underrepresented regions were examined by Sanger sequencing. The NGS approach was successfully established using cases with known sequence alterations. Depending on the initial clinical diagnosis, we identified likely causative mutations in 55% of retinitis pigmentosa and 80% of Bardet–Biedl or Usher syndrome cases. Seventy-one novel mutations in 40 genes were newly associated with RD. The genes USH2A, EYS, ABCA4, and RHO were more frequently affected than others. Occasionally, cases carried mutations in more than one RD-associated gene. In addition, we found possible dominant de-novo mutations in cases with sporadic RD, which implies consequences for counseling of patients and families. NGS-based mutation analyses are reliable and cost-efficient approaches in gene diagnostics of genetically heterogeneous diseases like RD.
Similar content being viewed by others
Introduction
Worldwide, >2 million patients are affected by monogenic forms of hereditary retinal dystrophies (RD). RD frequently leads to a significant impairment of the patients’ visual abilities and, depending on the subtype and course of the disease, may cause blindness. At least 20 diagnoses can be differentiated (for review see Berger et al.1). Although the forms of RD can be distinguished clinically, the disease progression is difficult to predict and may vary even within the same family.
A striking feature of the disease is its genetic heterogeneity, best illustrated by the fact that mutations in >100 genes are known to cause the different forms of RD.1 The majority of RDs can be subdivided into diseases primarily leading to rod- or cone- or generalized photoreceptor degenerations. The most frequent rod-dominated RD subtype is retinitis pigmentosa (RP), where mutations in >50 genes cause a similar phenotype featuring night blindness and mid-peripheral vision loss.2 Hereditary retinal diseases affecting mainly the central vision include cone- or cone-rod dystrophies as well as macular dystrophies. These forms of RD have been associated with >30 genes. Moreover, mutations in >10 genes cause juvenile blindness with generalized photoreceptor dystrophy (ie Leber congenital amaurosis (LCA)). A variety of syndromic forms such as Usher- and Bardet–Biedl syndrome feature RD and have been associated with >20 genes (for reference also see RetNet, https://sph.uth.edu/retnet/).
RD can be inherited as autosomal recessive (ar) or autosomal dominant (ad), as well as X-linked (XL), or mitochondrial traits. Nevertheless, the majority of cases are sporadic. Identifying the genetic cause of the patients’ disease is crucial for genetic counseling of patients and families, and is a prerequisite for any form of genotype-based therapies. However, the enormous genetic heterogeneity in RD makes attempts to identify causative mutations a challenging task. Standard procedures in molecular gene diagnostics of RD include techniques like Sanger sequencing or array-based mutation screenings. The arrayed primer extension (APEX) technology is specifically designed to analyze previously described mutations and thus detects pathogenic sequence alterations in only 10–20% of RP or congenital stationary night blindness cases.3, 4 Similar technologies applied for Stargardt disease or LCA patients resulted in higher detection rates.5, 6 In contrast to the APEX technology, Sanger and microarray-based sequencing approaches identify also novel base changes.7, 8 Approaches to apply Sanger sequencing to all known disease genes implicated in a subtype of RD, namely RP, result in a detection rate of 50–60% among the autosomal dominant RP cases, however, only at the expense of an enormous effort in time and costs for the analysis of hundreds of exons.2, 9 Recently, studies applying next generation sequencing (NGS) techniques to analyze genetically precharacterized patient collections showed similar mutation detection rates for RP cases.10, 11, 12, 13
Though NGS is not yet routinely used in gene diagnostics of RD, this technology is capable of handling the tremendous genetic heterogeneity of these diseases through massively parallelizing the sequencing of all known RD-associated genes.12 In this report, we describe a diagnostic NGS pipeline to identify mutations in an unselected cohort of 170 patients with different forms of RD.
Materials and Methods
Please refer to the Supplementary material for additional sections of the Material and Methods.
Mutation definition
Herein, sequence alterations that were considered to be likely disease causing are denoted as mutations. We applied the following criteria to describe a likely pathogenic sequence change as ‘mutation’: (i) the sequence change has previously been documented to be pathogenic in the literature, (ii) it results in a shift of the open reading frame of the transcript, (iii) it is a nonsense mutation introducing a premature stop codon, (iv) it changes the canonical splice-site sequence, or (v) it causes an alteration of the deduced amino-acid sequence of the protein that is predicted to be damaging (according to MutationTaster or Alamut). In cases where an amino acid alteration or a splice defect can be assumed, we used prediction programs to further evaluate the putative pathogenicity of the sequence alteration (NetGene2 or Alamut). Of note, the frame-shifting mutations mostly constitute small deletions or insertions, although a few patients showed deletions of complete exons.
A case presenting a family history with affected members in at least two subsequent generations was considered to be solved when a heterozygous mutation was found in a gene that has previously been associated with dominant inheritance. However, one exception is made in these autosomal dominant families: an unknown missense change was not considered to be causative without verification of its cosegregation with the phenotype. In cases with presumed autosomal recessive inheritance, at least a single likely pathogenic mutation in combination with a second mutation in the same gene was considered to explain the disease of the patient.
Results
Patients
The analyzed patients (n=170) mostly originated from Germany and occasionally from neighboring countries including Switzerland, Austria and the United Kingdom. The majority of the patients had a well-defined clinical diagnosis, which included either RP, cone- or cone-rod dystrophy, macular dystrophy, LCA, Usher syndrome, or Bardet–Biedl syndrome. The referral of the patients’ sample included information on the putative underlying mode of inheritance. In 34 cases, samples from additional family members were used to verify segregation of the sequence variants identified in the index patient.
Next generation sequencing of diagnostic panels
Following quality control and fragmentation of the patient’s genomic DNA, we performed an in-solution enrichment and NGS of 105 genes (in total 1650 exons with ∼316 000 bp) in which mutations had previously been associated with RD (Figure 1). To facilitate the molecular gene diagnostics of the RD-associated genes bioinformatically, we defined subpanels that included all genes known to cause one of the following diseases: RP, cone- or cone-rod dystrophy, Stargardt disease or macular dystrophy, LCA, Usher syndrome, and Bardet–Biedl syndrome (Table 1). The category of RP was further subdivided into two panels (autosomal recessive/X-linked and autosomal dominant/X-linked genes). The search for mutations explaining the phenotype started by evaluating the clinically most relevant subpanel(s). If the case was not solved by this procedure, all additional subpanels were analyzed.
On average, the SOLiD sequencing and mapping resulted in a coverage of 783 reads per base pair (bp) and patient and showed 53.5% or 4703623 reads on target. Poorly covered genomic regions were bioinformatically identified when the coverage per nucleotide dropped below 10-fold. Approximately 5% of the bp fall into this category and were re-analyzed by Sanger sequencing to meet diagnostic criteria and to ensure a reliable analysis of disease-associated gene regions. In the largest subpanels, we frequently verified up to 30 exons. In three cases (nos. 154, 927 and 1309) the causative mutation(s) were detected only by the Sanger sequencing of underrepresented exons (Supplementary Table 1).
The variant calls generated by LifeScope or Bioscope software packages were annotated using a local copy of the Ensembl database,14 dbSNP and in-house variant databases. Variants were selected for further investigation, if the global minor allele frequency of the sequence change was <5% based on dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) and the Exome Variant Server (http://evs.gs.washington.edu/EVS/). Following this bioinformatic filtering, the variants were classified as missense, nonsense, splice-site (within 10 bp of splice acceptor or donor site), non-synonymous or synonymous near splice-site (within 2 bp at the beginning or end of the exon). A sequence change was excluded when found in >20% of all cases from the in-house database.
Verification of the diagnostic strategy
To verify the reliability of our NGS strategy, we tested three samples in which causative mutations and polymorphisms had previously been identified by Sanger sequencing (Supplementary Table 2). These cases were diagnosed as either autosomal recessive Stargardt disease, autosomal dominant macular dystrophy, or autosomal dominant RP. In addition to several polymorphic variants, four likely causative mutations locate to the genes ABCA4, RHO or PRPH2. In total, the three positive controls contained 32 previously detected sequence alterations distributed over several coding exons in 5 genes (Supplementary Table 2).
The NGS approach redetected all but one polymorphic variant. The single undetected sequence change was located in exon-flanking regions with low coverage, and thus was not annotated. Using the NGS approach, we were able to redetect ∼97% of the sequence variants previously found by Sanger sequencing. This value is in accordance with our previous observation that ∼5% of the gene regions (underrepresented in the NGS) should be verified by Sanger sequencing to ensure a reliable diagnostic screening for each patient sample.
Mutation identification in 170 unselected RD cases
We analyzed 170 patients with different forms of RD. The patients were not selected for specific clinical and genetic criteria.
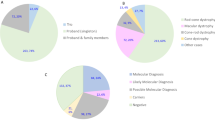
The majority of cases (n=111; 65%) were diagnosed with RP. The remaining patients were affected by different forms of syndromic RP (Usher syndrome: n=20, 12%; Bardet–Biedl syndrome: n=6, 3.5%), cone- or cone-rod dystrophies (n=18, 10.5%), macular dystrophies (n=10, 6%), LCA (n=4, 2%), or congenital stationary night blindness (n=1, 0.5%).
Mutations were identified in the majority of the analyzed cases (Supplementary Tables 1, 3 and 4). Among patients with a clinical diagnosis of RP, we detected mutations explaining the disease phenotype in 62 out of 112 cases. Thus, we obtained a diagnostic detection rate of 55% for RP. This value includes that 41% (12 out of 29) of the autosomal dominant and 60% (50 out of 82) of the autosomal recessive and sporadic RP cases were associated with mutations. Similar percentages of solved cases are found among the other monogenic forms of RD. Higher detection rates were obtained in cases with syndromic phenotypes. In 80% (21 out of 26) of the patients with Usher- and Bardet–Biedl syndrome, either homozygous or compound heterozygous mutations were identified.
We found 47 novel mutations in cases diagnosed with RP (Supplementary Tables 1, 3 and 4). In addition, the screening of patients with cone- and cone-rod dystrophy, macular dystrophy, and LCA revealed eight novel mutations. Among the phenotypes of Usher- and Bardet–Biedl syndrome, 16 new mutations were found. Altogether, the 170 analyzed cases carried 71 mutations that have not been described before and were newly associated with different forms of RD.
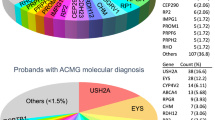
Approximately one-third of the detected mutations were missense changes (Figure 2). These were either known or predicted to be damaging. The categories of nonsense mutations and small deletions together account for ∼44% of the identified mutations. In addition, 12% of the variants fall into the category of splice-site mutations or intronic mutations and are likely to induce missplicing of the affected transcript. Of note, we found three cases with larger deletions covering single or multiple exons in Usher syndrome-associated genes (twice in USH2A and once in PCDH15; patients nos. 125, 459 and 1426).
In total, we detected mutations in 40 different RD genes (Figure 2). The prevalence of disease alleles among the RD genes was not equally distributed. Mutations in USH2A were most frequently identified and accounted for 23 cases. Mutations in EYS, ABCA4, and RHO explained eight, five and five cases, respectively. Yet, recurrent mutations were uncommon and most mutations were only found once or twice.
Wherever possible, the cosegregation of the mutations and the phenotype within the respective family was verified. This analysis was performed in 34 cases, although the number of additional family members available was often small and included mostly the parents and occasionally siblings (Supplementary Table 5). Segregation analysis was concordant in all but two cases, in which either the detected sequence alterations are not associated with the disease or incomplete penetrance modifies the expressivity of the phenotype (Supplementary Table 1 and Supplementary Table 5).
Unsolved cases
In 15 patients (9%), we did not detect sequence alterations that completely explain the disease phenotype. Supplementary Table 6 lists cases, in which we found ambiguous variants or unclear combinations of sequence changes.
Novel missense mutations that were bioinformatically predicted to be polymorphisms are frequently found in these unsolved cases. As additional family members were not available for segregation analyses, the pathogenicity of these sequence alterations could not be verified.
In order to apply a conservative interpretation of the pathogenicity of the detected mutations, novel amino acid exchanges that occurred in cases with dominant inheritance were categorized to be unclear (even if the sequence alteration was predicted to be damaging). Nevertheless, these mutations may be causative for the disease phenotype, but additional studies are required to verify this notion.
We also detected mutations at positions other than the canonical splice-site (first two or last two intronic nucleotide positions) that show a high potential to interfere with splicing. For example, mutations at position +5 of the splice donor site are frequently found to cause splice defects. This is supported by bioinformatic prediction programs that result in a significant reduction of the splice-site score. Nevertheless, further studies are required to evaluate the effect of these mutations on splicing. Consequently, these variants are categorized unclear.
Interestingly, several of these cases showed alterations not only in a single disease gene, but carried mutations in more than one gene. Supplementary Table 6 summarizes those cases where we did not find a clear correlation between the phenotype and the identified mutations within a single disease gene.
Accumulation of multiple mutations in a single patient
Among the solved sporadic or autosomal recessive cases, several patients carried, in addition to disease-associated mutations, alteration in other genes (Supplementary Table 7). Occasionally, these additional sequence changes were also classified to be mutations and occurred in RD genes associated with autosomal recessive inheritance. For example, in case no. 459 we found a homozygous frame-shifting deletion of exon 14 in USH2A that occurred together with heterozygous mutations in ABCA4 and RPE65. Both, the ABCA4 and RPE65 mutations have previously been described to be pathogenic.15, 16 Fourteen similar cases were identified (Supplementary Table 7), suggesting that occasionally patients may be affected by mutations in more than one causative gene.
Discussion
NGS is capable of sequencing all known RD-associated genes in parallel, generating millions of reads from preselected genomic regions. This is a clear advantage compared with conventional Sanger sequencing, where only a few hundred base pairs of a single sample can be analyzed per reaction. In addition, NGS technologies in combination with DNA bar coding enable the simultaneous analysis of several patient samples. Especially for genetically heterogeneous diseases like RD, high-throughput techniques are among the most promising and economic approaches to identify causative mutations.
We developed a diagnostic NGS pipeline that targets the genomic regions of 105 retinal disease-associated genes. For the specific purpose of molecular gene diagnostics, this strategy shows advantages compared with exome sequencing. Although economic reasons calculated on a cost per base pair are the least convincing, it clearly increases the number of patients analyzed per run and thus, the throughput of the diagnostic analysis pipeline. In addition, the costs of bioinformatic applications and data storage can be minimized. The reliability of the NGS data depends on the coverage per base pair. We reached an average coverage of ∼750-fold, whereas this value is usually significantly less in exome analyses. The applied technologies were able to improve the coverage across the target genes with only 5% of the base pairs having a coverage below 10-fold. This reduces the efforts of Sanger sequencing in the attempt to verify the less well covered regions. Last but not the least, generating exome data for diagnostic purposes raises ethic concerns on the sequencing of gene regions not associated with a specific diagnosis.
Our diagnostic NGS pipeline enabled the identification of mutations explaining the patients’ disease phenotype in 55–80%. The values vary depending on the initial clinical diagnosis. The NGS detection rates described herein were generated by analyses of 170 RD patient samples that were neither clinically nor genetically preselected, a circumstance that, to the best of our knowledge, has not been described before in RD. Another study applied a next generation sequencing (NGS) technology to a genetically pre-characterized cohort of 100 patients with simplex or autosomal recessive RP.11 The authors extrapolated from their data that in an unbiased cohort, a diagnostic yield of 47% might have been achieved. Audo et al10 found values of 57% in 17 pre-screened families where the majority of cases presented with RP and congenital stational night blindness. Shanks et al17 tested 36 patients and proposed a higher detection rate in the early onset cases. Interestingly, Neveling et al11 and Audo et al10 analyzed their patient samples on two NGS platforms different from the one described in this study, suggesting that the major NGS platforms yield similar mutation detection rates,18 even for a tremendously heterogeneous group of diseases like RP. Indeed, this observation is encouraging for attempts to use NGS as a diagnostic tool.12 However, extensive optimization and evaluation procedures are required for all NGS platforms to ensure a reliable and routine application of NGS technologies in diagnostics.
Several disease genes are associated with specific inheritance patterns and thus, the search for causative mutations may be guided by the family pedigree. Nevertheless, the majority of the 170 analyzed patient samples have been referred to us as being sporadic cases lacking any family history for RD. These cases are especially challenging for gene diagnostics and counseling. Most of them can be expected to follow an autosomal recessive mode of inheritance, but dominant de-novo mutation occur.19 This is also supported by our data, as we found several sporadic RD cases that show likely pathogenic mutations in genes that predominantly have been associated with autosomal dominant inheritance. Although additional studies are required to show the pathogenicity of these sequence alterations, at least nine patients with RP or cone-dominated diseases fall into this category (Supplementary Table 8). De-novo mutations were not identified in patients with syndromic forms of RD, likely because the corresponding genes are exclusively inherited in an autosomal recessive manner. Importantly, cases with suspected de-novo mutations may have an increased risk of having affected children, a serious problem for counseling of the patients with sporadic RD. In ∼10% (4 out of 39) of the sporadic RP cases, a putative de-novo mutation occurred in an autosomal dominantly inherited gene. We even found 5 out of 13 cases with potential de-novo mutations in cone-dominated diseases (Supplementary Table 8). Although it requires further data to reliably predict the rate of de-novo mutations among the sporadic cases, this observation should be considered during genetic counseling of patients. Of note, these mutations would have been overlooked by more targeted strategies to identify the causative mutations in autosomal recessively inherited genes.
An additional aspect relevant to the counseling of patients arose from cases who carry mutations associated with incomplete penetrance. In one of the analyzed cases (patient no. 534), a previously described missense mutation was detected in SNRNP200, a splice factor associated with autosomal dominant RP.20, 21 Not only the affected patient, but also the unaffected mother (determined by the patient history) carried the mutation, either suggesting incomplete penetrance or a non-causative relation of the mutation and the phenotype. As the same mutation has been published to cause the disease,20, 21 it seems likely that incomplete penetrance occurred in the family described herein. Interestingly, mutations in another splice factor gene, PRPF31, have also been associated with incomplete penetrance.22 A second case (patient no. 325), where incomplete penetrance might be newly described herein, is a cone-rod dystrophy patient carrying a heterozygous stop mutation in PROM1. The affected sister and the unaffected father also carried this mutation, suggesting that reduced penetrance occurred in the father. Nevertheless, it cannot be excluded that the phenotype in this family is explained by two recessive PROM1 mutations,23, 24 rather than a single dominant one.25, 26
Among the unsolved autosomal recessive or sporadic cases, amino acid alterations often were bioinformatically predicted to be polymorphisms. Published data was evaluated to verify these predictions. However, some of these sequence changes may also be mutations. Additional cases or functional tests are required to evaluate the disease-relevance of the individual changes. Furthermore, it cannot be excluded that deep intronic variants exist that were not detected due to the targeted enrichment of the exonic regions for NGS-based genetic testing. To identify a previously described pathogenic variant that is located deep in the intron 26 of CEP290 (c.2991+1655A>G),27 we generally performed Sanger sequencing of the genomic interval in patients with Leber congenital amaurosis. However, we found several sporadic or autosomal recessive cases that carry sequence alterations in one of the frequently affected genes (eg USH2A, EYS, ABCA4 or PDE6B), but lack a second clearly pathogenic variant. Of note, these genes are among the largest associated with RD. It will be interesting to perform whole-genome sequencing on these cases in order to analyze the regulatory or intronic gene regions for additional mutations. Alternatively, patient-derived cell lines might be screened for splice defects or expression level changes to test for functional consequences of pathogenic variants outside of the coding region.
Additional mutations cannot only occur in the same disease-associated gene. Sequence changes in different genes may have cumulative effects on the clinical presentation of the patients’ disease and thus, are considered to be genetic modifiers of the phenotype. At least two different types of modifier effects have been described in RD. Either two distinct phenotypes occur in the same patient or the progression of the disease is altered. Neveling et al11 described a patient affected by RP (caused by PDE6B mutations), in whom a modifying pathogenic sequence alteration in PRPH2 led to additional features of a macular degeneration. A similar case where Best disease and congenital stationary night blindness cosegregated in the same family was recently described.10 Furthermore, Poloschek et al28 characterized a family where modifier mutations in ABCA4 and ROM1 result in a cumulative effect worsening the macular degeneration phenotype caused by a PRPH2 mutation. In the present study, we also found several cases where modifier mutations might influence the phenotype, but additional clinical and genetic tests are required to verify this notion.
Together, the identification of mutations in heterogeneous diseases like RD is increasingly dependent on high-throughput sequencing technologies. We and others have demonstrated the diagnostic value of NGS platforms. In addition, these data suggest that the so-called monogenic retinal diseases are indeed influenced by additional genetic factors explaining, at least in parts, the enormous clinical variability seen among the patients.
References
Berger W, Kloeckener-Gruissem B, Neidhardt J : The molecular basis of human retinal and vitreoretinal diseases. Prog Retin Eye Res 2010; 29: 335–375.
Hartong DT, Berson EL, Dryja TP : Retinitis pigmentosa. Lancet 2006; 368: 1795–1809.
Avila-Fernandez A, Cantalapiedra D, Aller E et al: Mutation analysis of 272 Spanish families affected by autosomal recessive retinitis pigmentosa using a genotyping microarray. Mol Vis 2010; 16: 2550–2558.
Zeitz C, Labs S, Lorenz B et al: Genotyping microarray for CSNB-associated genes. Invest Ophthalmol Vis Sci 2009; 50: 5919–5926.
Zernant J, Kulm M, Dharmaraj S et al: Genotyping microarray (disease chip) for Leber congenital amaurosis: detection of modifier alleles. Invest Ophthalmol Vis Sci 2005; 46: 3052–3059.
Jaakson K, Zernant J, Kulm M et al: Genotyping microarray (gene chip) for the ABCR (ABCA4) gene. Hum Mutat 2003; 22: 395–403.
Gonzalez-del PM, Borrego S, Barragan I et al: Mutation screening of multiple genes in Spanish patients with autosomal recessive retinitis pigmentosa by targeted resequencing. PLoS One 2011; 6: e27894.
Clark GR, Crowe P, Muszynska D et al: Development of a diagnostic genetic test for simplex and autosomal recessive retinitis pigmentosa. Ophthalmology 2010; 117: 2169–2177.
Daiger SP, Sullivan LS, Bowne SJ et al: Targeted high-throughput DNA sequencing for gene discovery in retinitis pigmentosa. Adv Exp Med Biol 2010; 664: 325–331.
Audo I, Bujakowska KM, Leveillard T et al: Development and application of a next-generation-sequencing (NGS) approach to detect known and novel gene defects underlying retinal diseases. Orphanet J Rare Dis 2012; 7: 8.
Neveling K, Collin RW, Gilissen C et al: Next-generation genetic testing for retinitis pigmentosa. Hum Mutat 2012; 33: 963–972.
O'Sullivan J, Mullaney BG, Bhaskar SS et al: A paradigm shift in the delivery of services for diagnosis of inherited retinal disease. J Med Genet 2012; 49: 322–326.
Simpson DA, Clark GR, Alexander S, Silvestri G, Willoughby CE : Molecular diagnosis for heterogeneous genetic diseases with targeted high-throughput DNA sequencing applied to retinitis pigmentosa. J Med Genet 2011; 48: 145–151.
Grant JR, Arantes AS, Liao X, Stothard P : In-depth annotation of SNPs arising from resequencing projects using NGS-SNP. Bioinformatics 2011; 27: 2300–2301.
Lewis RA, Shroyer NF, Singh N et al: Genotype/phenotype analysis of a photoreceptor-specific ATP-binding cassette transporter gene, ABCR, in Stargardt disease. Am J Hum Genet 1999; 64: 422–434.
Morimura H, Fishman GA, Grover SA, Fulton AB, Berson EL, Dryja TP : Mutations in the RPE65 gene in patients with autosomal recessive retinitis pigmentosa or leber congenital amaurosis. Proc Natl Acad Sci USA 1998; 95: 3088–3093.
Shanks ME, Downes SM, Copley RR et al: Next-generation sequencing (NGS) as a diagnostic tool for retinal degeneration reveals a much higher detection rate in early-onset disease. Eur J Hum Genet 2012; 21: 274–280.
Benaglio P, Rivolta C : Ultra high throughput sequencing in human DNA variation detection: a comparative study on the NDUFA3-PRPF31 region. PLoS One 2010; 5: 9.
Vissers LE, de LJ, Gilissen C et al: A de novo paradigm for mental retardation. Nat Genet 2010; 42: 1109–1112.
Benaglio P, McGee TL, Capelli LP, Harper S, Berson EL, Rivolta C : Next generation sequencing of pooled samples reveals new SNRNP200 mutations associated with retinitis pigmentosa. Hum Mutat 2011; 32: E2246–E2258.
Zhao C, Bellur DL, Lu S et al: Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. Am J Hum Genet 2009; 85: 617–627.
Rivolta C, McGee TL, Rio FT, Jensen RV, Berson EL, Dryja TP : Variation in retinitis pigmentosa-11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum Mutat 2006; 27: 644–653.
Zhang Q, Zulfiqar F, Xiao X et al: Severe retinitis pigmentosa mapped to 4p15 and associated with a novel mutation in the PROM1 gene. Hum Genet 2007; 122: 293–299.
Permanyer J, Navarro R, Friedman J et al: Autosomal recessive retinitis pigmentosa with early macular affectation caused by premature truncation in PROM1. Invest Ophthalmol Vis Sci 2010; 51: 2656–2663.
Michaelides M, Gaillard MC, Escher P et al: The PROM1 mutation p.R373C causes an autosomal dominant bull's eye maculopathy associated with rod, rod-cone, and macular dystrophy. Invest Ophthalmol Vis Sci 2010; 51: 4771–4780.
Yang Z, Chen Y, Lillo C et al: Mutant prominin 1 found in patients with macular degeneration disrupts photoreceptor disk morphogenesis in mice. J Clin Invest 2008; 118: 2908–2916.
den Hollander AI, Koenekoop RK, Yzer S et al: Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet 2006; 79: 556–561.
Poloschek CM, Bach M, Lagreze WA et al: ABCA4 and ROM1: implications for modification of the PRPH2-associated macular dystrophy phenotype. Invest Ophthalmol Vis Sci 2010; 51: 4253–4265.
Acknowledgements
We would like to thank all patients for participating in this study. We are grateful for technical assistance from Esther Glaus and Mariana Wittmer. We thank Detlef Boehm for his help in setting up the first versions of the panel. He was employed by CeGaT during that time. This work was in part supported by a grant of the BMBF (01GM1108A to BW and SK) and the Forschungskredit of the University of Zurich (to JN).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Glöckle, N., Kohl, S., Mohr, J. et al. Panel-based next generation sequencing as a reliable and efficient technique to detect mutations in unselected patients with retinal dystrophies. Eur J Hum Genet 22, 99–104 (2014). https://doi.org/10.1038/ejhg.2013.72
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.72
Keywords
This article is cited by
-
Unearthing FLVCR1a: tracing the path to a vital cellular transporter
Cellular and Molecular Life Sciences (2024)
-
A novel homozygous missense substitution p.Thr313Ile in the PDE6B gene underlies autosomal recessive retinitis pigmentosa in a consanguineous Pakistani family
BMC Ophthalmology (2023)
-
Genetic tool used to diagnose achromatopsia: first case report from India
Egyptian Journal of Medical Human Genetics (2023)
-
No strong evidence to date for an association between RIMS1 and retinal dystrophy
Documenta Ophthalmologica (2023)
-
Autosomal dominant optic atrophy caused by six novel pathogenic OPA1 variants and genotype–phenotype correlation analysis
BMC Ophthalmology (2022)