Abstract
Cholera is endemic to many countries. Recent major outbreaks of cholera have prompted World Health Organization to recommend oral cholera vaccination as a public-health strategy. Variation in percentage of seroconversion upon cholera vaccination has been recorded across populations. Vaccine-induced responses are influenced by host genetic differences. We have investigated association between single-nucleotide polymorphic (SNP) loci in and around 296 immunologically relevant genes and total anti-lipopolysaccharide (LPS) antibody response to a killed whole-cell vaccine, comprising LPS from multiple strains of Vibrio cholerae. Titers derived from standard vibriocidal assays were also analyzed to gain further insights on validated SNP associations. Vaccination was administered to 1000 individuals drawn from India. Data on two independent random subsets, each comprising ∼500 vaccinees, were used for discovery of genomic associations and validation, respectively. Significant associations of four SNPs and haplotypes in three genes (MARCO, TNFAIP3 and CXCL12) with AR were discovered and validated, of which two in TNFAIP3 and CXCL12 were also significantly associated with immunity (fourfold increase in vibriocidal titers). CXCL12 is a neutrophil and lymphocyte chemoattractant that is upregulated in response to V. cholerae infection. LPS in the vaccine possibly provides signals that mimic those of the live bacterium. TNFAIP3 promotes intestinal epithelial barrier integrity and provides tight junction protein regulation; possible requirements for adequate response to the vaccine. LPS is a potent activator of innate immune responses and a ligand of MARCO. Variants in this gene have been found to be associated with LPS response, but not with high vibriocidal titer level.
Similar content being viewed by others
Introduction
Major outbreaks of cholera, leading to several thousand deaths, in Zimbabwe in 2009 and in Haiti in 2010, prompted the World Health Organization to emphasize the administration of oral cholera vaccination.1, 2, 3, 4 Cholera is caused by toxicogenic strains of the Gram-negative bacterium Vibrio cholerae. Moderate to severe watery diarrhea, that appears within 1–5 days of ingestion of contaminated food or water, rapidly leads to dehydration and, if left untreated, to death. It is a significant cause of mortality in developing countries,5 and is still endemic in many countries, including India.6 Unfortunately, 13% of the world’s population still lacks access to safe water.7
V. cholerae, has two major serogroups – O1 (serotypes Inaba and Ogawa) and O139 – that are responsible for most cholera cases and differ in the lipopolysachharide (LPS) structures of the outer membrane.8 Immunity to cholera can be acquired.9 WHO advocates vaccination for cholera.10 The efficacy of currently available vaccines varies across populations; protective efficacy is unlikely to last more than 3 years.11 A single-dose cholera vaccine that induces long-term immunity for those at risk remains an unfulfilled goal.12 Persons of blood group O are more susceptible to cholera than non-O blood group persons.13, 14, 15 However, the association between efficacy of cholera vaccines and O blood group has been inconsistent–negative in Bangladesh,13 positive in Indonesia16 and no association in India.17 Inter-individual variability in immune response to vaccines impedes the control of infectious diseases. Primary failure, as assessed by postvaccination antibody levels, of vaccines is a problem; 2–10% for measles vaccine,18, 19 5–20% for hepatitis B vaccine20, 21 and 23–40% for typhoid vaccine.22, 23 Data are unavailable for cholera vaccines. Twin studies have shown high heritability for antibody response (AR) to measles (89%), mumps (39%), rubella (46%), hepatitis B (61–77%), oral polio (60%), tetanus (44–64%), diphtheria (49%), Haemophilus influenzae type B (Hib) (51%) and other vaccines.24 Polymorphic variants in HLA and other genes–such as, IL1B, IL4R, IL6, IL10 and TNF–are associated or linked with AR to several vaccines.18, 25, 26, 27, 28, 29, 30 Although measles, mumps and rubella vaccines are all attenuated-live vaccines that are administered simultaneously, the responses induced by these vaccines are influenced differently by host genetics.24 Vietnam is the only country in the world with large cholera-endemic tracts where an oral vaccine for cholera has been widely used for a long time (since 1997).31 Considerable variation in response among vaccinees was noted in a clinical trial of this vaccine31 conducted in a non-endemic area of Vietnam (Son La province), although >90% of vaccinees seroconverted. However, in respect of the same vaccine, only ∼50% of vaccinees seroconverted in the endemic area of West Bengal, India.17
Vaccine response is not wholly determined by the genetic make-up of vaccine recipients, but also by the nature of microbial antigens and adjuvants.32 Knowledge of key immunogenetic associations can be used to design a vaccine that circumvents immunogenetic restrictions.33 Insights into the impact of genetic variation in vaccinees on AR has been obtained for some vaccines.24, 34, 35 In this study, we have investigated, for the first time, whether variation in the genomic backgrounds of vaccinees contributed to variation in immunological response to the killed whole-cell bivalent oral cholera vaccine, which has been introduced in India in 2010.
Methods
Study participants
Individuals (n=1000), unrelated at least to the first-cousin level based on family history report, aged 14 years or older, inhabiting a socio-economically depressed locality of Kolkata (formerly, Calcutta), India, were recruited using a statistical sampling design,34 with institutional ethical approval and written individual informed consent. Exclusions included: (a) pregnant and lactating women and (b) based on self-report, individuals who had ever been diagnosed with cholera or had suffered from diarrhea or vomiting during the week preceding recruitment. The sample of 948 vaccinees, whose data passed all the quality checks (explained later), was randomly split into two half-samples (HSs); one used for association-discovery and the other for association-validation.
Vaccination and collection of blood samples
With approval of the Drug Controller General of India, a two-dose vaccine (see online Supplementary Table S1 for composition) procured from Shantha Biotechnics, Hyderabad, India, orally administered 14 days apart, was used. The vaccine was stored and transported at 4–8 °C before administration. From each vaccinee, a blood sample was collected immediately before vaccination (Day 0 sample) and 28 days postvaccination (Day 28 sample).
Assessment of antibody level
Antibody level was assessed using two different methods as described below.
Total anti-LPS AR assay
Serum anti-V. cholerae (Inaba 569) LPS–that is, total anti-LPS antibody–was assayed using the Bio-Plex (Bio Rad, Hercules, CA, USA) platform (hereafter referred to as the ‘Bioplex assay’). V. cholerae O1 (Inaba 569) LPS (List Biological, Cat. No.231) was conjugated to Bio Rad beads (COOH (042)) by Solulink (San Diego, CA, USA). Day 0 and Day 28 human sera were incubated with LPS beads with anti-V. cholerae LPS antibody and responses were detected using a cocktail of anti-human IgG-PE, IgA-PE and IgM-PE. A pooled serum with a high anti-LPS IgG/IgA/IgM response was used to create a standard curve that was assigned an arbitrary value of anti-LPS Ig ELISA units (EU)/ml. Responses (Day 0 and Day 28) in individual subjects were compared with the standard curve to calculate anti-LPS Ig EU/ml.
Vibriocidal assay
This assay was performed with V. cholerae O1 Inaba (OS-418), Ogawa (MAK757) and uncapsulated O139 (MO-10T4) strains using sera collected during pre- and postvaccine trials following published methods.36 Commercially available guinea-pig serum (Rockland, Gilbertsville, PA, USA) was used as a source of complement. The sera (100 μl) were added to 100 μl of PBS in the first well to give twofold dilution and the subsequent dilutions were made reciprocally up to 4800. Reference rabbit antiserum against V. cholerae O1 Inaba, Ogawa and O139 were included in each set of assay as controls. In every batch of assay, sera obtained from a healthy volunteer and a high-titer antiserum obtained from one of the volunteers in this study were included as negative and positive controls, respectively.
Candidate genes and SNPs
We selected 296 autosomal genes from immunological pathways as candidates for this association study. Two thousand four hundred and eighty-eight SNPs from these genes (Supplementary Table S2 online), including about 2-kb upstream and and 1-kb downstream regions, were chosen from the HapMap database (www.hapmap.org) using a statistical protocol37 that maximized ‘informativeness’ of selected SNPs. Individuals belonging to the first HS were genotyped at all the 2488 loci. Genotypes at those loci that showed statistically significant association with AR in the first HS were determined in individuals belonging to the second HS for validation of the discovered associations. Genotyping was done using the Illumina iSCAN (San Diego, CA, USA) platform using Golden Gate assays following protocols recommended by the manufacturer.
Statistical analysis
AR to vaccination was measured as the ratio (fold-change) of Day 28 postvaccination to prevaccination (Day 0) antibody levels (EU/ml). Individuals on whom AR could not be properly assayed (5%) were removed from analyses. Curation of genotype data included removal of (a) loci with <90% call rate (2% of loci), (b) non-polymorphic loci with MAF<0.05 (12.7% of loci with call rate >90%), (c) loci significantly deviant from HWE (1% of polymorphic loci), and (d) individuals with <90% of loci with valid genotype calls (0.7%). After data-curation, 948 individuals and 2036 SNPs were included for further analysis. Analysis of variance (ANOVA) was performed to test equality of mean values of lnAR (loge-transformation was used to induce Normality) among subgroups of vaccinees. To avoid detection of spurious associations arising from systematic ancestry differences, population stratification analysis was performed using Eigenstrat.38 Instead of using an arbitrary threshold to classify a vaccinee as a ‘responder’ or a ‘non-responder’, we sorted the vaccinees by ascending order of their AR and grouped the vaccinees into five pentile (20 percentile) groups (PGs). It is expected that the frequency of an allele that is significantly associated with response should increase (or decrease) from low responders to high responders. Detection of allelic association was done by estimating the correlation coefficient (r) between the MAF at each locus and mean AR of vaccinees belonging to the (PGs) and testing H0: r=0 using a permutation test. The false discovery rate procedure39 was used to identify statistically significant values of r at the 5% level of significance. As correction for multiple testing when loci are in significant linkage disequilibrium (LD) remains a major statistical problem,40, 41 for single-locus association analysis, we circumvented the problem by selecting from each gene only the highly informative and minimally associated SNPs.37 Four hundred and sixty-nine SNPs were thus selected; data on the remaining loci were kept aside for haplotype association analysis. Each significant allelic association was revalidated by confirming that the null hypothesis that the mean logAR values among the genotype classes are equal was rejected, using ANOVA. The standard regression procedure was used to estimate effect size (slope, β) of each significantly associated SNP. SD of effect size was estimated by bootstrapping 100 times; that is, by randomly choosing the appropriate number of samples with replacement from the original data and estimating β from the bootstrap sample and then calculating the empirical SD of the 100 estimates of β. Haplotypes were inferred using Phase.42 Haplotype association analysis was performed using a permutation algorithm similar to that for single SNP markers.
Results
Demographic characteristics of vaccinees
No statistically significant differences in the demographic characteristics of the vaccinees (Table 1) were noted between the association discovery and validation HSs in respect of age (mean ages were 34.24±0.60 years and 34.36±0.58 years; p=0.883), proportions of vaccinees belonging to the two genders (p=0.602) or the two religious groups (p=0.21).
Inter-individual variation in pre- and post-vaccination antibody levels and AR
Large inter-individual variations of pre- and postvaccination antibody levels and also of AR to vaccination (fold-change) were noted (Table 2 and Supplementary Table S3 online). Mean values between the two HSs were not significantly different (Supplementary Table S3). The correlation between baseline and Day 28 antibody levels is positive and moderate (0.449), but significant (p<0.0001).
Antibody response
AR-estimates obtained from the two separate assays were significantly (p<0.0001) correlated (Table 3). As, by its very nature, the vibriocidal assay is less quantitative than the bioplex assay, in further analyses, unless specifically mentioned, AR as measured by the bioplex assay was used. Mean values of lnAR for the two HSs did not differ significantly (p=0.564) and were, 1.128±0.038 and 1.160±0.039, respectively. None of the possible covariates–religion, age and gender–had a statistically significant effect on lnAR, either individually or interactively (p-values for main effects and interactions were all >0.05). Therefore, all data were pooled in subsequent analyses.
Population stratification
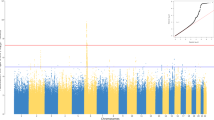
No pair of vaccinees in the first HS shared >90% of the 2036 loci with identical genotypes; i.e., there was no evidence of cryptic relatedness among the vaccinees. There was also no evidence of population stratification as all individuals formed a single cluster in the scatterplot of the first three principal components (Figure 1).
Antibody levels and response in vaccinees belonging to the five PGs
Mean values of antibody levels and response were nearly equal for each PG between the HSs (Supplementary Table S4 online), but differed significantly (P<0.05) among the PGs (Table 4). (Mean values of fold-change based on the vibriocidal assay were consistently higher than those based on the bioplex assay.) Mean AR increased almost linearly from PG 1 through 4, after which there was a steep increase (Figure 2), which was also observed for values based on vibriocidal assay. The steep increase of mean AR in PG-5 is primarily because of the presence of some individuals who exhibited very high AR.
Association results: discovery and validation
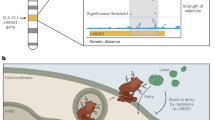
In the discovery HS, correlations between mean AR and MAFs in PGs were found to be statistically significant at 159 SNP loci on 93 genes (of the 296 genes considered in this study; Supplementary Table S5 online). The associations of these 159 SNPs could be validated in HS-2 only for seven SNPs that are located in and around five genes (rs17180481 and rs17180600 in the flanking region and intron, respectively, of MARCO (2q14.2); rs598493 in an intron of TNFAIP3 (6q23.3); rs3012694 and rs266087 in introns of RFX3 (9p24.2); rs266087 in an intron of CXCL12 (10q11.21); and rs6567272 in an intron of TNFRSF11A (18q22.1)). Mean AR for vaccinees belonging to the three genotypes for each of these seven SNP loci differed significantly for only the four SNPs in and around MARCO, TNFAIP3 and CXCL12 (rs17180481: p=0.023, effect size=1.8±0.3; rs17180600: p=0.026, effect size=1.6±0.3; rs598493: p=0.001, effect size=3.9±0.8; rs266087: p=0.014, effect size=2.4±0.6), but not for the remaining three. The MAF values in PGs are provided in Figure 3 for these four SNPs.
Haplotype association
For these three genes, data on additional SNPs, which were in significant LD with the SNPs included in the association analysis, were used to reconstruct haplotypes and estimate haplotype frequencies among the vaccinees. Figure 4 provides, for MARCO, TNFAIP3 and CXCL12, the frequency of the haplotype that showed the maximum association with AR by PG. The strength of association (i.e., slope of the line) with AR increased significantly for haplotypes compared with the single SNPs in these genes, indicating that these three genes are important modulators of AR to the vaccine.
Frequencies of haplotypes containing the minor allele at the significantly associated SNP loci in the pooled sample PGs. (a) Haplotype GGAGCG of CXCL12, (b) Haplotype GAAGCG of CXCL12, (c) Haplotype GAAGC of TNFAIP3, (d) Haplotype AA of MARCO. Boldfaced and underlined alleles are those the minor alleles at the corresponding loci. (Another haplotype–GGAGC of TNFAIP3–had lower frequencies than those of the haplotype presented in this figure and was absent in three PGs.).
Concordance between anti-LPS antibody and vibriocidal responses
For the four loci that were validated to be significantly associated with AR, the proportions of individuals who elicited fourfold or higher titer increase in the vibriocidal assay (that is likely to be suggestive of protective immunity,43, 44 although true protective immunity must be ascertained by vaccine challenge studies) significantly differed among the three genotypes at the locus only for two (rs598493 in TNFAIP3; p=0.001 and rs266087 in CXCL12; p=0.014) of these four loci (Figure 5). Thus, although MARCO was found to be significantly associated with AR, it was not found to be associated with high vibriocidal titer levels.
Discussion
WHO has recommended that cholera vaccination be provided in many geographical regions.2 Protective immune response to V. cholerae that colonizes the intestinal mucosal surface without invasion of enterocytes45 is believed to reside at the mucosal surface.44 Orally administered vaccines maximize the intestinal secretory AR, and a long-lasting memory so that the primed intestinal immune system can rapidly respond to a subsequent exposure to antigen.46 Serum vibriocidal activity, measured by a bactericidal assay requiring the fixation of complement by antibody that is bound specifically to vibrios, is used extensively as a marker of immunity to V. cholerae, but has limitations.47, 48 On the basis of recent reccommendations49 to measure LPS-response in multiple immunoglobulin isotypes and results50, 51 which indicate that levels of all immunoglobulin isotypes (IgG, IgM and IgA) determine the nature and extent of protection afforded by oral cholera vaccination, we have used the total anti-LPS AR and also the traditional vibriocidal assay.
Clinical trials of many oral vaccines have underscored the importance of understanding the reasons for variability in immune response to oral vaccines,52 and the identification of genetic factors in the host linked to vaccine immunogenicity. It has been emphasized that such identification may result in adoption of ‘inverse of personalized medicine’ approaches to design vaccines to overcome barriers in nonresponding populations.53 Susceptibility to cholera is significantly associated with a marker in the promoter region of LPLUNC1 (rs11906665),54 an innate immunity gene that likely has a role in modulating host inflammatory responses to V. cholerae infection.55 We have discovered and validated that three genes–MARCO, TNFAIP3 and CXCL12–has significant roles in modulating AR. However, MARCO was not significantly associated with increased vibriocidal titers. A single cell layer of the mucosal epithelium essentially regulates intestinal homeostasis. This physical barrier is a prominent part of the innate immune system with damage-healing and infection-limiting properties. CXCL12 is a homeostatic or constitutive chemokine that has a key role in healing by stimulating the activation of laminin-specific integrins.56 V. cholerae infection produces strong inflammatory manifestations.57 CXCL12 is a neutrophil and lymphocyte chemoattractant that is upregulated in response to V. cholerae infection, resulting in increased recruitment of polymorphonuclear leukocytes to the site of infection.58 The LPS in the cholera vaccine possibly provides signals that mimic those of the live bacterium, without actually causing disease. The TNF-induced protein 3 (TNFAIP3) is an ubiquitin-modifying enzyme and an essential negative regulator of inflammation, through the inhibition of NF-κB activation and prevention of TLR-mediated responses.59 Tight junctions between intestinal epithelial cells mediate the permeability of the intestinal barrier. TNFAIP3 has a role in promoting intestinal epithelial barrier integrity and in enabling maintenance of intestinal homeostasis through tight junction protein regulation;60 possible requirements for adequate response to the whole-cell killed cholera vaccine used in this study. TNFAIP3 has also been implicated in genome-wide studies of susceptibility of several autoimmune disorders.61 MARCO is a type II transmembrane protein of the class A scavenger receptor family.62 LPS, the major component of the vaccine used in this study, is a very potent activator of innate immune responses. LPS stimulation upregulates surface expression of MARCO in a dose- and time-dependent fashion.63 MARCO is upregulated in bacterial infections in macrophages of most tissues; variants in this gene have crucial roles in bacterial binding.64 No SNP in MARCO was found to be associated with high vibriocidal titer, possibly because different immunoglobulin isotypes are associated with anti-LPS and vibriocidal responses.65
The inferences of this study carried out in a cholera-endemic area, where many vaccinees possibly have prior exposure to the pathogen and consequently have high-baseline antibody levels (Table 2), may require validation in a non-endemic area. We, however, emphasize that the vaccine-response studies are more relevant and meaningful from a public-health viewpoint if conducted in endemic areas. In individuals with prior exposure to V. cholerae, the administered vaccine possibly acted as a booster, implying that individuals with high-baseline antibody levels are expected to show a low response (fold-change) to the vaccine, while individuals with low-baseline antibody levels are expected to show a high response. Although this feature was weakly observed (Table 2), the Day 0 and Day 28 antibody levels were significantly (P<0.0001) positively correlated (r=0.449). In this study we have used quantitative level of AR. Therefore, genomic associations with antibody fold-change detected in this study are unlikely to be affected by baseline antibody levels. However, it may be tricky is to relate the association between the level of AR and a SNP in a gene of the immune system with the level of immunological protection. In an endemic area, individuals with the lowest AR may be the best protected, while in a non-endemic area the reverse may be true. Although these issues pertaining to long-term protection remain unresolved, the genes that have shown significant association with AR are all biologically relevant. Functional studies on the markers found to be associated with AR need to be carried out, which together with the findings on LPLUNC1,54, 55 may provide a deep mechanistic understanding of the mode of action of the whole-cell killed oral cholera vaccine.
References
World Health Organization: Cholera: global surveillance summary, 2008. Weekly Epid Rec 2009; 84: 309–324.
World Health Organization: Meeting of the Strategic Advisory Group of Experts on immunization October 2009–conclusions and recommendations. Weekly Epid Rec 2009; 50: 526–528.
Date KA, Vicari A, Hyde TB et al: Considerations for oral cholera vaccine use during outbreak after earthquake in Haiti 2010-2011. Emerg Infect Dis 2011; 17: 2105–2112.
Tappero JW, Tauxe-Emer RV : Lessons learned during public health response to cholera epidemic in haiti and the dominican republic. Emerg Infect Dis 2011; 17: 2087–2093.
Institute of Medicine: New Vaccine Development Establishing Priorities Volume II Diseases of Importance in Developing Countries. Washington DC: National Academy Press, 1986.
World Health Organization:: Cholera, 2010. Weekly Epid Rec 2011; 86: 325–340.
Ryan ET : The cholera pandemic still with us after half a century Time to rethink. PLoS Negl Trop Dis 2011; 5: e1003.
Nandy RK, Sengupta TK, Mukhopadhyay S, Ghosh AC : A comparative study of the properties of Vibrio cholera 0139 01 and other non-01 strains. J Med Microbiol 1995; 42: 251–257.
Levine MM, Black RE, Clements ML, Cisneros L, Nalin DR, Young CR : Duration of infection-derived immunity to cholera. J Infect Dis 1981; 143: 818–820.
World Health Organization: Global Task Force on Cholera Control. Cholera Vaccines A new public health tool? Report of a WHO meeting 10–11 December 2002 Geneva Switzerland World Health Organization 2002, pp 14.
Sinclair D, Abba K, Zaman K, Qadri F, Graves PM : Oral vaccines for preventing cholera. Cochrane Database Syst Rev 2011; (Issue 3): Art No CD008603.
Provenzano D, Kovác P, Wade WF : The ABCs (Antibody B cells and Carbohydrate epitopes) of cholera immunity considerations for an improved vaccine. Microbiol Immunol 2006; 50: 899–927.
Clemens JD, Sack DA, Harris JR et al: ABO blood groups and cholera new observations on specificity of risk and modification of vaccine efficacy. J Infect Dis 1989; 159: 770–773.
Glass RI, Holmgren J, Haley CE et al: Predisposition for cholera of individuals with O blood groupPossible evolutionary significance. Am J Epidemiol 1985; 121: 791–796.
Harris JB, Khan AI, LaRocque RC et al: Blood group immunity and risk of infection with Vibrio cholerae in an area of endemicity. Infect Immun 2005; 73: 7422–7427.
Lagos R, Avendaño A, Prado V et al: Attenuated live cholera vaccine strain CVD 103-HgR elicits significantly higher serum vibriocidal antibody titres in persons of blood group O. Infect Immun 1995; 63: 707–709.
Ramamurthy T, Wagener D, Chowdhury G, Majumder PP : A large study on immunological response to a whole-cell killed oral cholera vaccine reveals that there are significant geographical differences in response and that O blood group individuals do not elicit a higher response. Clin Vaccine Immunol 2010; 17: 1232–1237.
Poland GA : Variability in immune response to pathogens using measles vaccine to probe immunogenetic determinants of response. Am J Hum Genet 1998; 62: 215–220.
Poland GA, Jacobson RM, Colbourne SA et al: Measles antibody seroprevalence rates among immunized Inuit Innu and Caucasian subjects. Vaccine 1999; 17: 1525–1531.
Milich DR, Leroux-Roels GG : Immunogenetics of the response to HBsAg vaccination. Autoimmun Rev 2003; 2: 248–257.
Zuckerman JN : Nonresponse to hepatitis B vaccines and the kinetics of anti-HBs production. J Med Virol 1996; 50: 283–288.
Gupta D, Faridi MM, Aggarwal A, Kaur I : Seroprevalence of anti-Vi antibodies and immunogenicity of Typhim Vi vaccine in children. Hum Vaccine 2008; 4: 305–308.
Sur D, Ochiai RL, Bhattacharya SK et al: A cluster-randomized effectiveness trial of Vi typhoid vaccine in India. N Engl J Med 2009; 361: 403–405.
Kimman TG, Vandebriel RJ, Hoebee B : Genetic variation in the response to vaccination. Community Genet 2007; 10: 201–217.
Kruskall MS, Alper CA, Awdeh Z, Yunis EJ, Marcus-Bagley D : The immune response to hepatitis B vaccine in humans inheritance patterns in families. J Exp Med 1992; 175: 495–502.
Gelder CM, Lambkin R, Hart KW et al: Associations between human leukocyte antigens and nonresponsiveness to influenza vaccine. J Infect Dis 2002; 185: 114–117.
Poland GA, Ovsyannikova IG, Jacobson RM : Immunogenetics of seasonal influenza vaccine response. Vaccine 2008; 26: D35–D40.
Poland GA, Ovsyannikova IG, Jacobson RM : Vaccine immunogenetics bedside to bench to population. Vaccine 2008; 26: 6183–6188.
van Eden W, de Bries RRP, Stanford JL, Rook GAW : HLADR3 associated genetic control of response to multiple skin tests with new tuberculins. Clin Exp Immunol 1983; 52: 287–292.
Wang C, Tang J, Song W, Lobashevsky E, Wilson CM, Kaslow RA : HLA and cytokine gene polymorphisms are independently associated with responses to hepatitis B vaccination. Hepatology 2004; 39: 978–988.
Anh DD, Canh do G, Lopez AL et al: Safety and immunogenicity of a reformulated Vietnamese bivalent killed whole-cell oral cholera vaccine in adults. Vaccine 2007; 25: 1149–1155.
Pulendran B, Kumar P, Cutler CW, Mohamadzadeh M, Van Dyke T, Banchereau J : Lipopolysaccharides from distinct pathogens induce different classes of immune responses in vivo. J Immunol 2001; 167: 5067–5076.
Jin P, Wang E : Polymorphism in clinical immunology—from HLA typing to immunogenetic profiling. J Transl Med 2003; 1: 8.
Majumder PP, Staats HF, Sarkar-Roy N et al: Genetic determinants of immune-response to a polysaccharide vaccine for typhoid. HUGO J 2009; 3: 17–30.
Ovsyannikova IG, Haralambieva IH, Vierkant RA, Pankratz VS, Jacobson RM, Poland GA : The role of polymorphisms in Toll-like receptors and their associated intracellular signaling genes in measles vaccine immunity. Hum Genet 2011; 130: 547–561.
Benenson AS, Saad A, Mosley WH : Serological studies in cholera 2 The vibriocidal antibody response of cholera patients determined by a microtechnique. Bull World Health Organ 1968; 38: 277–285.
Sarkar-Roy N, Mondal D, Bhattacharya P, Majumder P : A novel statistical algorithm for enhancing the utility of HapMap data to design genomic association studies in non-HapMap populations. Int J Data Mining Bioinf 2011; 5: 706–716.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D : Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909.
Benjamini Y, Hochberg Y : Controlling the false discovery rate a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B Methodological 1995; 57: 289–300.
Conneely KN, Boehnke M : So many correlated tests so little time! Rapid adjustment of P values for multiple correlated tests. Am J Hum Genet 2007; 81: 1158–1168.
Han B, Kang HM, Eskin E : Rapid and accurate multiple testing correction and power estimation for millions of correlated markers. PLoS Genet 2009; 5: e1000456.
Stephens M, Smith N, Donnelly P : A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 2001; 68: 978–989.
Eko FO, Schukovskaya T, Lotzmanova EY et al: Evaluation of the protective efficacy of Vibrio cholerae ghost VCG; candidate vaccines in rabbits. Vaccine 2003; 21: 3663–3674.
Tarique AA, Kalsy A, Arifuzzaman M et al: Transcutaneous immunization with a Vibrio cholerae O1 Ogawa synthetic hexasaccharide conjugate following oral whole-cell cholera vaccination boosts vibriocidal responses and induces protective immunity in mice. Clin Vaccine Immunol 2012; 19: 594–602.
Sack DA, Sack RB, Nair GB, Siddique AK : Cholera. Lancet 2004; 363: 223–233.
Kaper JB, Morris JG, Levine MM : Cholera. Clin Microbiol Rev 1995; 8: 48–86.
Clemens JD, van Loon F, Sack DA et al: Field trial of oral cholera vaccines in Bangladesh serum vibriocidal and antitoxic antibodies as markers of the risk of cholera. J Infect Dis 1991; 163: 1235–1242.
Saha D, LaRocque RC, Khan AI et al: Incomplete correlation of serum vibriocidal antibody titer with protection from Vibrio cholerae infection in urban Bangladesh. J Infect Dis 2004; 189: 2318–2322.
Losonsky GA, Yunyongying J, Lim V et al: Factors influencing secondary vibriocidal immune responses Relevance for understanding immunity to cholera. Infect Immun 1996; 64: 10–15.
Alam MM, Riyadh MA, Fatema K et al: Antigen-specific memory B-cell responses in Bangladeshi adults after one- or two-dose oral killed cholera vaccination and comparison with responses in patients with naturally. Clin Vaccine Immunol 2011; 18: 844–850.
Leung DT, Rahman MA, Mohasin M et al: Memory B Cell and Other Immune Responses in Children Receiving Two Doses of an Oral Killed Cholera Vaccine Compared to Responses following Natural Cholera Infection in Bangladesh. Clin Vaccine Immunol 2012; 19: 690–698.
Levine MM : Immunogenicity and efficacy of oral vaccines in developing countries lessons from a live cholera vaccine. BMC Biol 2010; 8: 129.
Serazin AC, Shackelton LA, Wilson C, Bhan MK : Improving the performance of enteric vaccines in the developing world. Nat Immunol 2010; 11: 769–773.
LaRocque RC, Sabeti P, Duggal P et al: A variant in long palate lung and nasal epithelium clone 1 is associated with cholera in a Bangladeshi population. Genes Immun 2009; 10: 267–272.
Shin OS, Uddin T, Citorik R et al: LPLUNC1 modulates innate immune responses to Vibrio cholera. J Infect Dis 2011; 204: 1349–1357.
Agle KA, Vongsa RA, Dwinell MB : Chemokine stimulation promotes enterocyte migration through laminin-specific integrins. Am J Physiol Gastrointest Liver Physiol 2011; 301: G968–G980.
Ou G, Rompikuntal PK, Bitar A et al: Vibrio cholerae cytolysin causes an inflammatory response in human intestinal epithelial cells that is modulated by the prtv protease. PLoS ONE 2009; 4: e7806.
Poggi A, Carosio R, Fenoglio D et al: Migration of Vδ1 and Vδ2T cells in response to CXCR3 and CXCR4 ligands in healthy donors and HIV-1–infected patients competition by HIV-1 Tat. Blood 2003; 103: 2205–2213.
Jennner RG, Young RA : Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol 2005; 3: 281–294.
Kolodziej LE, Lodolce JP, Chang JE et al: TNFAIP2 maintains intestinal barrier function and supports epithelial cell tight junctions. PLoS One 2011; 6: e26352.
Fung EY, Smyth DJ, Howson JM et al: Analysis of 17 autoimmune disease-associated variants in type 1 diabetes identifies 6q23/TNFAIP3 as a susceptibility locus. Genes Immunity 2009; 10: 188–191.
Kraal G, van der Laan LJW, Elomaa O, Tryggvason K : The macrophage receptor MARCO. Microbes Infect 2000; 2: 313–316.
Van der Laan LJW, Dopp EA, Haworth R et al: Regulation and functional involvement of macrophage scavenger receptor MARCO in clearance of bacteria in vivo. J Immunol 1999; 162: 939–947.
Sankala M, Brännström A, Schulthess T et al: Characterization of Recombinant Soluble Macrophage Scavenger Receptor MARCO. J Biol Chem 2002; 277: 33378–33385.
Alam MM, Riyadh MA, Fatema K et al: Antigen-specific memory B-cell responses in bangladeshi adults after one- or two-dose oral killed cholera vaccination and comparison with responses in patients with naturally acquired cholera. Clin Vac Immuol 2011; 18: 844–850.
Acknowledgements
Financial support for this work was provided by U.S. National Institute of Allergy and Infectious Diseases, National Institutes of Health Contract HHSN200400067C. The vaccine was used in this study with approval of the Drug Controller General of India, and the entire study was approved by the Health Ministry Monitoring Committee, Government of India; we are grateful to them for the approvals. We are grateful to members of CpG and TCGA for their expert help in the conduct of this project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Majumder, P., Sarkar-Roy, N., Staats, H. et al. Genomic correlates of variability in immune response to an oral cholera vaccine. Eur J Hum Genet 21, 1000–1006 (2013). https://doi.org/10.1038/ejhg.2012.278
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2012.278





 ) and validation (
) and validation ( ) half-samples, and in the pooled (
) half-samples, and in the pooled ( ) sample (±SD in the pooled sample) in PGs. (a) rs266087 (CXCL12), (b) rs598493 (TNFAIP3), (c) rs17180481 (MARCO) and (d) rs17180600 (MARCO).
) sample (±SD in the pooled sample) in PGs. (a) rs266087 (CXCL12), (b) rs598493 (TNFAIP3), (c) rs17180481 (MARCO) and (d) rs17180600 (MARCO).
