Abstract
Although over 200 pathogenic mitochondrial DNA (mtDNA) mutations have been reported to date, determining the genetic aetiology of many cases of mitochondrial disease is still not straightforward. Here, we describe the investigations undertaken to uncover the underlying molecular defect(s) in two unrelated Caucasian patients with suspected mtDNA disease, who presented with similar symptoms of myopathy, deafness, neurodevelopmental delay, epilepsy, marked fatigue and, in one case, retinal degeneration. Histochemical and biochemical evidence of mitochondrial respiratory chain deficiency was observed in the patient muscle biopsies and both patients were discovered to harbour a novel heteroplasmic mitochondrial tRNA (mt-tRNA)Ser(AGY) (MTTS2) mutation (m.12264C>T and m.12261T>C, respectively). Clear segregation of the m.12261T>C mutation with the biochemical defect, as demonstrated by single-fibre radioactive RFLP, confirmed the pathogenicity of this novel variant in patient 2. However, unusually high levels of m.12264C>T mutation within both COX-positive (98.4±1.5%) and COX-deficient (98.2 ±2.1%) fibres in patient 1 necessitated further functional investigations to prove its pathogenicity. Northern blot analysis demonstrated the detrimental effect of the m.12264C>T mutation on mt-tRNASer(AGY) stability, ultimately resulting in decreased steady-state levels of fully assembled complexes I and IV, as shown by blue-native polyacrylamide gel electrophoresis. Our findings expand the spectrum of pathogenic mutations associated with the MTTS2 gene and highlight MTTS2 mutations as an important cause of retinal and syndromic auditory impairment.
Similar content being viewed by others
Introduction
Mitochondrial DNA (mtDNA) mutations are estimated to affect at least 1 in 10 000 people in the UK1 and more than 200 pathogenic mtDNA mutations have been reported to date (MITOMAP: A Human Mitochondrial Genome Database, http://www.mitomap.org/MITOMAP). These mutations are located across the 37 genes of the mitochondrial genome; however, the vast majority reside in just 5–10% of the mtDNA, in the 22 mitochondrial tRNA (mt-tRNA) genes.
Mt-tRNA mutations are associated with a wide range of clinical disorders, from multisystemic syndromes including MERRF (myoclonic epilepsy and ragged-red fibres) and MELAS (mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes) to isolated organ-specific diseases such as deafness, retinopathy, myopathy or cardiomyopathy.2, 3 Mutations are unevenly distributed across the MTT genes, with over a third residing in just three of these genes: MTTL1, MTTI and MTTK. In contrast, MTTS2 represents one of the least affected mt-tRNA genes. To date, only two MTTS2 mutations have been reported, for which sufficient evidence exists to support their classification as ‘definitely pathogenic’ according to recently revised canonical criteria.4, 5 These mutations are m.12258C>A, which can lead to progressive deafness and either diabetes mellitus6 or retinitis pigmentosa,7 and m.12262C>A, which has been shown to cause progressive mitochondrial myopathy, deafness and sporadic seizures.8 A further three MTTS2 mutations have been described: m.12207G>A, which is associated with a MELAS-like phenotype,9 m.12236G>A, which is linked to non-syndromic hearing impairment,10 and m.12246C>A, which is associated with chronic intestinal pseudo-obstruction with myopathy and ophthalmoplegia.11 However, due to a lack of functional investigations (eg, single-fibre analysis, trans-mitochondrial cybrid studies), their pathogenicity has not yet been unequivocally proven.
Here, we report on two novel aminoacyl acceptor stem MTTS2 mutations that have been identified in two unrelated Caucasian patients, both displaying symptoms of myopathy, deafness, neurodevelopmental delay, epilepsy, marked fatigue and, in one case, retinitis pigmentosa. The histochemical, biochemical and molecular genetic data confirming the pathogenicity of these novel mtDNA substitutions are described.
Materials and methods
Case reports
This study had the relevant institutional ethical approval and complied with the Declaration of Helsinki.
Patient 1
Patient 1 is the first child of healthy, non-consanguineous Caucasian parents and was born following a normal pregnancy and delivery. Although early neurodevelopment was thought to be normal, by 12 months she was exhibiting signs of psychomotor retardation. Independent walking was delayed until 24 months and her gait remains unsteady. A mild, progressive generalised myopathy was associated with profound fatigue. Early signs of language acquisition were not sustained and she gradually lost speech. Feeding difficulties and failure to thrive were prominent early features, only partly resolved by the insertion of a gastrostomy, and she remains short and thin (>2.5 SD below mean for height and weight). A diagnosis of atypical autism with severe learning difficulties was made at the age of 4 years when cranial MRI was reported as normal. Epilepsy, in the form of absence seizures, developed later at age 10 years, when sensorineural hearing impairment was also noted.
Patient 2
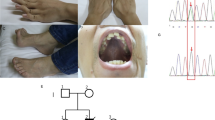
Patient 2 was born to non-consanguineous Caucasian parents and although pregnancy had been complicated by polyhydramnios, the delivery, at 41 weeks gestation, was normal. A pectus excavatum was noted during an early examination, but no other abnormalities were identified. By 5 years, dysmorphic features including neck webbing and micrognathia were evident, suggesting Noonan syndrome, along with significant psychomotor delay and learning difficulties. The patient developed a sensorineural hearing loss and was deaf by age 11 years. The first of many complex partial seizures occurred at age 10 years and had been preceded by 12 months of staring spells; an electroencephalogram confirmed a diagnosis of epilepsy, but cranial MRI was normal. Although there were no feeding difficulties, height and weight remained below the 5th percentile. A progressively severe proximal myopathy was associated with decreased muscle bulk and marked fatigue following minimal exertion. Balance appeared poor, but Romberg's test was negative and there was no upper-limb dysmetria. Cardiac evaluation revealed occasional ectopic beats and abnormal T waves on ECG, but no evidence of cardiomyopathy. Eye examination identified reduced visual acuity, pale optic discs and a mottled pigmentary retinopathy. The patient complained of light sensitivity, but not of night blindness. Electroretinogram demonstrated severe photoreceptor dysfunction. At 29 years of age, bilateral dense cataracts caused further visual obscuration necessitating surgery. The patient's mother also had pectus excavatum, progressive exercise intolerance, migraine and night-blindness with pigmentary retinopathy. Two sisters had poor night vision and hypothyroidism, while a brother had pectus excavatum but no other abnormalities.
Mitochondrial ATP production, respiratory chain enzyme activities and morphological analysis
Tibialis anterior muscle specimens were obtained from patient 1 in 1999 and 2009, at 3 and 13 years of age, respectively, using a percutaneous conchotome biopsy procedure. Mitochondrial ATP production rate (MAPR) and respiratory chain enzyme activities were determined from isolated mitochondria as previously described.12 Standard techniques were applied for routine and enzyme histochemical staining of cryostat sections13 and for electron microscopy.14 Respiratory chain activities in isolated mitochondria from vastus lateralis muscle from patient 2 were measured as described previously.15, 16
Cell culture
Cultured skin fibroblasts from patient 1 and her mother were maintained as previously described.17
Nucleic acid extraction
Total DNA was extracted from skeletal muscle, blood, cultured skin fibroblasts and urine and buccal epithelia by standard procedures. Total DNA from individual COX-deficient and COX-positive muscle fibres, isolated by laser-microcapture, was obtained as described elsewhere.18 Total RNA was extracted from cultured primary skin fibroblasts with Trizol reagent (Life Technologies, Paisley, UK) according to manufacturer's instructions.
MtDNA sequencing
The entire mtDNA sequence was determined from total DNA isolated from fibroblasts from patient 1 and from skeletal muscle from patient 2. MtDNA was amplified by PCR in 28 (patient 1) or 36 (patient 2) overlapping M13-tailed fragments and amplicons were sequenced with BigDye Terminator cycle sequencing chemistries (Applied Biosystems, Warrington, UK) as described previously (Taylor et al19 and Tuppen et al,20 respectively). Sequence data were compared with the revised Cambridge reference sequence for human mtDNA (GenBank NC_012920.1).
Last hot cycle PCR restriction fragment length polymorphism analysis (PCR-RFLP)
Levels of mutant m.12261T>C and m.12264C>T mtDNA were determined by last hot cycle PCR-RFLP as previously described17 with the modifications detailed in Table 1. Levels of mutant mtDNA were calculated as a percentage of total mtDNA.
High-resolution northern blot analysis
High-resolution northern blot analysis of total fibroblast RNA (1–3 μg) or synthetic in vitro transcribed mt-tRNASer(AGY) (100pg) was performed as previously described.17 Human mt-tRNALeu(UUR) and mt-tRNASer(AGY) probes were generated using the primer pairs L3200 (nucleotide positions 3200–3219) 5′-TATACCCACACCCACCCAAG-3′ and H3353 (3353–3334) 5′-GCGATTAGAATGGGTACAAT-3′, and M13-tailed L12201 (12 201–12 223) 5′-tgtaaaacgacggccagtTTTACCGAGAAAGCTCACAAGA-3′ and M13-tailed H12270 (12 270–12 248) 5′-caggaaacagctatgaccAAAGTTGAGAAAGCCATGTTGTT-3′ (M13-tails shown in lower case), respectively. The radioactive signal of the mt-tRNASer(AGY) probe was normalised to that of the mt-tRNALeu(UUR) probe for each sample and ratios were expressed relative to an age-matched control.
Synthetic vector cloning
Template plasmids were constructed for m.12264C>T mutant and wild-type human MTTS2 using synthetic DNA fragments containing the class III promoter of the T7 RNA polymerase directly upstream of the mt-tRNASer(AGY) gene. These were synthesised by PCR using two overlapping primers; the forward primer 5′-TAATACGACTCACTATAGAGAAAGCTCACAAGAACTGCTAACTCATGCCCCC-3′ paired with either the wild-type or mutant reverse primer 5′-TG/AAGAAAGCCATGTTGTTAGACATGGGGGCATGAGTTAGCAGT-3′ (sequence differences are highlighted in bold: wild-type/mutant). These synthetic DNAs were cloned into the Srf I restriction site in the pPCR-Script Amp SK(+) cloning vector (Stratagene, Stockport, UK). Amplification of the ligated vector was achieved by transformation of XL10-Gold Kan ultracompetent cells. Plasmid DNA was extracted using the QIAprep Spin Miniprep kit (Qiagen, Crawley, UK) and sequenced using universal M13 primers.
In vitro transcription
Templates for T7 RNA polymerase transcription were generated from the extracted plasmid DNA by PCR, using the forward primer, 5′-TAATACGACTCACTATAGAGAAAGC-3′, paired with one of the following reverse primers: wild-type with (bold)/without 3′-CCA, 5′-TGG/TGAGAAAGCCATGTTGTTAGAC-3′, or mutant with (bold)/without 3′-CCA, 5′-TGG/TAAGAAAGCCATGTTGTTAGAC-3′. In vitro transcription was performed using the Ambion Megascript 7 kit according to the manufacturer's protocol.
Blue native-polyacrylamide gel electrophoresis (BN-PAGE)
Mitochondria isolated from the 2009 patient 1 muscle biopsy were analysed by BN-PAGE. Briefly, 20 μg mitochondria were pelleted, resuspended in 23 μl N-dodecyl β-D-maltoside buffer (1% N-dodecyl β-D-maltoside, 1.5 M 6-aminocaproic acid, 7 mM Bis-Tris, 2 mM EDTA, pH 7.0) and incubated for 15 min on ice. Following centrifugation at 20 000 g for 10 min at 4°C, supernatants were mixed with 7 μl sample buffer (750 mM 6-aminocaproic acid, 5% Coomassie Blue G250) and loaded onto a 5–15% gradient BN gel. Gel preparation and electrophoresis were carried out as described.21
Statistical analysis
Statistically significant differences in mutant mtDNA levels between COX-positive and COX-deficient muscle fibres were determined by the non-parametric Mann–Whitney U test. A P-value≤0.05 was considered statistically significant.
Results
Histochemistry and histology
Sequential COX-SDH histochemistry of skeletal muscle biopsies collected from patient 1 in 1999 (data not shown) and 2009 (Figure 1a) revealed 20–30% COX-deficient muscle fibres; <2% COX-deficient fibres were observed in the biopsy from the patient's mother (Figure 1b). No typical ragged red fibres (RRF) were identified in the 2009 patient biopsy, although modified Gomori trichrome staining did reveal accentuated peripheral red staining indicating a subsarcolemmal accumulation of mitochondria in occasional fibres (Figure 1c). Electron microscopy of the 2009 biopsy identified abnormal mitochondria with dense inclusions (Figure 1d).
Skeletal muscle histochemistry and histology. (a, b) COX-SDH staining of the 2009 skeletal muscle biopsy from patient 1 and mother, respectively. Blue-staining fibres indicate reduced COX activity. (c) Gomori trichrome staining of the 2009 patient 1 skeletal muscle biopsy. (d) Electron micrograph of the 2009 patient 1 skeletal muscle biopsy. Arrows indicate the presence of dense inclusions. Magnification x 24 000. (e– h) Staining of skeletal muscle from patient 2 for COX, SDH, Gomori trichrome and NADH-tetrazolium reductase, respectively.
Histochemical analysis of serial sections of skeletal muscle from patient 2 revealed approximately 6% COX-deficient fibres (Figure 1e) with increased SDH enzyme activity in subsarcolemmal areas (Figure 1f). Modified Gomori trichrome staining confirmed the presence of RRF in both COX-positive and COX-deficient fibres (Figure 1g). Both strong and weak NADH-TR-stained RRF were also observed (Figure 1h).
Biochemical results
Respiratory chain enzyme analysis of mitochondria isolated from the 2009 muscle biopsy sample from patient 1 revealed a marked reduction in the activities of complexes I and IV, with near normal activity of complex II (Figure 2a). A two-fold increase in citrate synthase activity suggests mitochondrial proliferation (Figure 2b). A substantial decrease in ATP production was noted for almost all substrates tested, most evidently with the complex I-dependent substrate combinations glutamate+malate and pyruvate+malate (Figure 2c). Interestingly, with succinate alone, a higher than normal MAPR was measured, a pattern previously observed in patients with isolated complex I deficiency.22 A biopsy from the patient's mother showed normal respiratory chain biochemistry results (data not shown). Biochemical analysis of patient 2 muscle mitochondria revealed a mild, isolated complex I deficiency (Figure 2d).
Biochemical analysis of skeletal muscle biopsies. (a) Respiratory chain enzyme activities in skeletal muscle mitochondria from patient 1 and controls (n=15, 5–60 years; mean±SD). I, NADH-coenzyme Q reductase; I+III, NADH-cytochrome c reductase; II, succinate-coenzyme Q reductase; II+III, succinate-cytochrome c reductase; IV, cytochrome c oxidase. Respiratory chain enzyme activities were determined as units/unit citrate synthase activity in the mitochondrial suspension. (b) Citrate synthase (CS) activity in skeletal muscle from patient 1. CS was determined as units/kg muscle. (c) Mitochondrial ATP production rate (MAPR) with substrates glutamate+succinate (G+S), glutamate+malate (G+M), TMPD+ascorbate (T+A), pyruvate+malate (P+A), palmitoyl-L-carnitine+malate (PC+M), succinate+rotenone (S+R) and succinate alone (S) determined as units/unit citrate synthase in isolated muscle mitochondria from patient 1 and controls. (d) Respiratory chain and citrate synthase activities in skeletal muscle mitochondria from patient 2 and controls (mean±SD). I, NADH-coenzyme Q reductase; II, succinate-coenzyme Q reductase; III, coenzyme Q-cytochrome c reductase; IV, cytochrome c oxidase; V, ATPase. Complexes III and IV were determined as first order rate constant/mg protein/min; all remaining activities were calculated as nmol/min/mg protein. All patient enzyme activities are expressed as a percentage of control mean.
Molecular genetic investigations
Complete sequencing of the mitochondrial genome in fibroblasts (patient 1) or skeletal muscle (patient 2) revealed novel substitutions within the MTTS2 gene in each patient, namely m.12264C>T (patient 1) and m.12261T>C (patient 2) (Figure 3a). Both transitions reside at well-conserved positions in the aminoacyl acceptor stem of mt-tRNASer(AGY) and disrupt Watson–Crick base-pairings (Figures 3b and c).
Novel MTTS2 substitutions, m.12264C>T and m.12262T>C. (a) Sequence electropherograms illustrating the transitions m.12264C>T (patient 1) and m.12261T>C (patient 2). (b) Mt-tRNASer(AGY) with the locations of the m.12264C>T and m.12261T>C substitutions highlighted. (c) Alignment of mt-tRNASer(AGY) aminoacyl acceptor (a.a.) stem sequences from Human (Homo sapiens, GenBank accession number NC_012920.1), Gorilla (Gorilla gorilla, NC_001645), Chimpanzee (Pan troglodytes, NC_001643), Cow (Bos taurus, NC_006853), Mouse (Mus musculus, NC_005089), Rat (Rattus norvegicus, NC_001665), Frog (Xenopus laevis, NC_001573), Yeast (Saccharomyces cerevisiae, NC_001224) and Fruit fly (Drosophila melanogaster, NC_001709). Locations of the m.12264C>T and m.12261T>C sequence variants are highlighted (boxes) along with their corresponding Watson–Crick base-pairing partners.
Quantitative analysis of the m.12264C>T and m.12261T>C substitutions
In order to ascertain the pathogenicity of the novel sequence variants, the levels of the m.12264C>T and m.12261T>C transitions were assessed by last hot cycle PCR-RFLP analysis in multiple tissues from patients 1 and 2 and their maternal ancestors. Mutant mtDNA levels were also measured in blood from patient 2's siblings. Very high levels of the m.12264C>T substitution (≥93%) were observed in patient 1 blood, urine and cultured skin fibroblasts, with near homoplasmic levels in skeletal muscle (Figure 4a). Single-fibre analyses surprisingly showed markedly high levels of mutant m.12664C>T mtDNA in both COX-positive (98.4±1.5%; n=11) and COX-deficient fibres (98.2±2.1%; n=11) (Figure 4b). Lower levels of mutant mtDNA, ranging from 9 to 36%, were found in tissues from the patient's mother (Figure 4a).
Quantitation of m.12264C>T and m.12261T>C transitions. (a) Last hot cycle PCR-RFLP analysis of the m.12264C>T sequence variant in blood (B), urine (U), cultured skin fibroblasts (F) and muscle (M) from patient 1 and her mother. Wild-type PCR products harbour one MaeIII restriction site, which cuts the amplicon into two fragments of 158 and 24 bp. Mutant amplicons contain two recognition sites, which produce three fragments upon MaeIII digest (136, 24 and 22 bp). Fragments were separated on a 12% non-denaturing PAGE gel. UN, uncut PCR amplicon; C, wild-type control. Percentage mutant load in each sample is indicated at the bottom of each lane. Fragment sizes (bp) are shown on the left. (b) Last hot cycle PCR-RFLP analysis of the m.12264C>T transition in single COX-positive (n=11) and COX-deficient (n=11) skeletal muscle fibres from the 2009 patient 1 biopsy. (c) Last hot cycle PCR-RFLP analysis of the m.12261T>C sequence variant in patient 2 muscle (M) and in blood (B) and urinary (U) and buccal (Bu) epithelia from patient 2 and his mother and grandmother. Wild-type PCR products harbour one MnlI restriction site, which cuts the amplicon into two fragments of 192 and 56 bp. Mutant amplicons contain two recognition sites, which produce three fragments upon MnlI digest (141, 56 and 51 bp). Fragments were separated on a 12% non-denaturing PAGE gel. UN, uncut PCR amplicon; C, wild-type control. Percentage mutant load in each sample is indicated at the bottom of each lane. Fragment sizes (bp) are shown on the left. (d) Last hot cycle PCR-RFLP analysis of the m.12261T>C sequence variant in single COX-positive (n=11) and COX-deficient (n=10) fibres from the patient 2 skeletal muscle biopsy.
Assessment of tissues from patient 2 revealed high levels of mutant m.12261T>C mtDNA in skeletal muscle (94%) but lower levels in urine (78%), buccal epithelial cells (15%) and blood (13%) (Figure 4c). Single-fibre analyses confirmed segregation of the substitution with the biochemical defect as significantly (P<0.0006, Mann–Whitney U test) higher levels of the m.12261T>C mutation were detected in COX-deficient fibres (97.1±1.1%; n=10) relative to COX-positive fibres (85.8±16.6%; n=11) (Figure 4d). Urine, buccal and blood samples from the patient's mother and maternal grandmother exhibited very low levels of the m.12261T>C transition, <3% (Figure 4c). No evidence of mutant mtDNA was detected in blood from patient 2's two brothers and two sisters (data not shown).
Functional investigations of the m.12264C>T substitution
In view of the near homoplasmic levels of the m.12264C>T transition in patient 1, functional consequences of the MTTS2 substitution were also investigated. Firstly, the steady-state levels of the mt-tRNA were assessed by high-resolution northern blot in cultured skin fibroblasts from the patient and her mother. Following standardisation to mt-tRNALeu(UUR) levels, a 70% reduction in mature mt-tRNASer(AGY) steady-state levels was determined in the patient's cells relative to age-matched controls (Figure 5a). Interestingly, a seemingly smaller subspecies of mt-tRNASer(AGY) was also detected in the patient's cells. Comparison with in vitro synthesised mt-tRNASer(AGY) molecules with and without 3′-CCA confirmed the subspecies was not an immature form of the mt-tRNA (Figure 5b). Further investigation of the identity of the mt-tRNASer(AGY) subspecies by tRNA circularisation and cloning failed to characterise this smaller molecule (data not shown). Neither a change in mt-tRNASer(AGY) steady-state levels nor a smaller subspecies were observed in the maternal fibroblasts.
Functional investigation of the m.12264C>T substitution. (a) High-resolution northern blot analysis of total RNA (1 μg) from cultured skin fibroblasts from patient 1 (P1), her mother (M) and paediatric (C1 and C2) and adult (C3 and C4) controls. Samples were separated on a 13%, 8 M urea-denaturing polyacrylamide gel, electroblotted onto membranes and hybridised with α32P-radiolabelled probes specific for mt-tRNALeu(UUR) and mt-tRNASer(AGY) transcripts. Mt-tRNASer(AGY) radioactive signals, normalised to those of their counterpart mt-tRNALeu(UUR), are expressed relative to the corresponding age-matched control C1 or C4. (b) High-resolution northern blot analysis of total RNA (3 μg) from cultured skin fibroblasts from patient 1 (P1) and in vitro synthesised mt-tRNASer(AGY) molecules (100pg) with (+) and without (−) 3′-terminus CCA addition. Membranes were probed exclusively for mt-tRNASer(AGY). (c) One-dimensional BN-PAGE (5–15%) of isolated mitochondria (20 μg) from patient 1 (P1) and control (C) muscle. Complexes I–V are identified (left).
Secondly, the expression and assembly of the mitochondrial OXPHOS complexes were assessed by 1D Blue-Native PAGE of isolated mitochondria from patient 1 muscle. A marked decrease in the amount of fully-assembled complex I was noted, as well as a less severe depletion in complex IV levels (Figure 5c).
Discussion
We describe two unrelated Caucasian patients, both underweight and of short stature, who presented with similar symptoms of myopathy, deafness, neurodevelopmental delay, epilepsy, marked fatigue and, in one case, retinitis pigmentosa, a form of genetic retinal degeneration or dystrophy. A suspected underlying mitochondrial respiratory chain disorder in each case was confirmed by the demonstration of OXPHOS defects in skeletal muscle biopsies, and primary mtDNA aetiologies were indicated by mosaic patterns of COX/SDH histochemical staining. Whole mitochondrial genome sequencing revealed two closely-residing novel substitutions within the aminoacyl acceptor stem of the MTTS2 gene.
To be considered pathogenic, mt-tRNA mutations must fulfil a number of accepted criteria.4, 5, 23, 24 Accordingly, neither the m.12264C>T nor the m.12261T>C sequence variants are recognised polymorphisms, both are located at well-conserved positions within the mt-tRNASer(AGY) aminoacyl acceptor stem and both are predicted to disrupt Watson–Crick base pairings at these sites with structural and subsequent functional implications for the affected mt-tRNA. Furthermore, last hot cycle PCR-RFLP analyses have demonstrated the two transitions to be heteroplasmic across a range of tissues and have highlighted a clear segregation of the substitutions with the disease in the two families. An additional critical indicator of pathogenicity for heteroplasmic mt-tRNA mutations is segregation of the mutation with the biochemical defect.4 Single-fibre analysis of muscle from patient 2 revealed a significant association between m.12261T>C mutant load and COX-deficiency and application of the recently revised canonical pathogenicity scoring system to m.12261T>C confirmed the mutation to be ‘definitely pathogenic’, with a score of 13 out of a possible 20 points.5 In contrast, near homoplasmic levels of the m.12264C>T substitution were found in both COX-positive and COX-deficient fibres from patient 1, and on the basis of these findings the variant could only be classed as ‘possibly pathogenic’ (score: 10/20). Further investigations of the functional effects of this transition on mt-tRNASer(AGY) were therefore warranted24 and the observation of a substantial reduction in steady-state levels of the affected mt-tRNA provided the necessary evidence for definite pathogenicity, as confirmed by a revised score of 15/20.5
Blue-Native PAGE analysis of skeletal muscle from patient 1 provided further insight into the potential functional consequences of the m.12264C>T mutation. Decreased levels of fully assembled complexes I and IV imply the mutation may impair mitochondrial protein synthesis. A reduction in mt-tRNASer(AGY) steady-state levels and the concomitant observation of a possible mt-tRNASer(AGY) subspecies by northern blot analysis suggest the m.12264C>T mutation may achieve this effect by destabilising the molecule, thereby targeting the mutated mt-tRNA for premature degradation or preventing correct 5′- and/or 3′-end processing of the molecule. In the event of the mutation impacting mt-tRNA maturation, an effect more profound than impairment of the 3′-CCA tail addition might be expected, given the estimated 10 nucleotide size difference between the mature mt-tRNASer(AGY) and the observed smaller molecule. Two mutations have previously been reported at nucleotide position 72 of MTTL1 (m.3303C>T) and MTTK (m.8363G>A). However, the varied functional consequences of these two mutations (m.3303C>T: reduced 3′-end processing and CCA addition25 but no impact on aminoacylation;26 m.8363G>A: unaffected mt-tRNA stability but impaired aminoacylation and de novo mitochondrial protein synthesis27) preclude us from extrapolating to the mutation reported here. Thus, further investigations will be necessary in order to fully understand the functional significance of the m.12264C>T mutation.
Observations of extremely high threshold levels for heteroplasmic mt-tRNA mutations are not usual and reports of mt-tRNA mutations reaching near homoplasmy within one generation are also not common, although we have previously described the rapid segregation of the MTTE m.14709T>C mutation to near universal tissue homoplasmy in three separate members of the same family,28 undoubtedly a result of an unfortunate segregation of affected mtDNA during oogenesis or early development. Mosaic patterns of COX-deficiency have previously been reported for pathogenic homoplasmic mt-tRNA mutations29, 30 and their phenotypic expression is considered to be dependent on a complex interaction with environmental and/or nuclear genetic factors.31, 32 The m.12264C>T mutation is obviously mild in nature and must require accumulation to near homoplasmic levels before a disruption of the electron transport is observed. Furthermore, factors such as fibre type and/or absolute wild-type mtDNA copy number may also be explanations for heterogeneous COX activity in skeletal muscle.
Four different ‘definitely pathogenic’ MTTS2 mutations, including the two described here, have now been reported.6, 7, 8 Interestingly, all four (m.12258C>A, m.12261T>C, m.12262C>A and m.12264C>T) are located within the 3′-end of the aminoacyl acceptor stem of mt-tRNASer(AGY), whereas a fifth ‘probably pathogenic’ MTTS2 mutation, m.12207G>A, has been identified within the 5′ end of this stem.9 This clustering of mutations within MTTS2 suggests that an intact aminoacyl acceptor stem is of crucial importance for maintaining the unusual secondary structure of mt-tRNASer(AGY), which lacks a DHU loop, and for preserving the correct function of the molecule. Although these pathogenic MTTS2 mutations are associated with a variety of clinical presentations including diabetes mellitus, myopathy, neurodevelopmental delay, encephalopathy and seizures, it is interesting to note that all four are also linked to deafness,6, 7, 8 and, like the vast majority of deafness-associated mt-tRNA mutations,33 both m.12264C>T and m.12261T>C were shown here to exhibit a high threshold for pathogenicity. Two additional MTTS2 sequence variants have also been linked to auditory symptoms; m.12236G>A has been associated with hearing loss10 and m.12224C>T, associated with haplogroup D4, has been found to modulate the penetrance of hearing loss associated with the MT-RNR1 mutation, m.1555A>G.34 Similar to MTTS1,35 the MTTS2 gene is emerging as an important hot spot for deafness-associated mutations. Notably, this is also the second report of an association between MTTS2 mutation and retinitis pigmentosa.
The specific phenotypic manifestation of MTTS mutations in the cochlea is intriguing and thus far remains unexplained. Several hypotheses have been proposed. One possibility may be a cochlea-specific reduction in the transcription of MTTS genes, thereby rendering them more susceptible to mutations than other mitochondrial genes.35 Alternatively, Fischel-Ghodsian et al36 have hypothesised that cochlea-specific isoforms or splice-variants of mitochondrial RNA-processing or translation proteins may be particularly sensitive to mutant mt-tRNAs, with resulting quantitative of qualitative modifications to the protein products. The inaccessibility of the cochlea, for in vivo sampling, will make it difficult to prove either of these hypotheses.
In summary, our data confirm the pathogenicity of two novel MTTS2 substitutions and extend both the clinical phenotype and genetic aetiology of mitochondrial respiratory chain disease, highlighting the emergence of MTTS2 mutations as an important cause of auditory impairment and retinitis pigmentosa.
Accession codes
Change history
19 July 2012
This article has been corrected since Advance Online Publication and a corrigendum is also printed in this issue
References
Schaefer AM, McFarland R, Blakely EL et al: Prevalence of mitochondrial DNA disease in adults. Ann Neurol 2008; 63: 35–39.
Anitori R, Manning K, Quan F et al: Contrasting phenotypes in three patients with novel mutations in mitochondrial tRNA genes. Mol Genet Metab 2005; 84: 176–188.
Scaglia F, Wong LJ : Human mitochondrial transfer RNAs: role of pathogenic mutation in disease. Muscle Nerve 2008; 37: 150–171.
McFarland R, Elson JL, Taylor RW, Howell N, Turnbull DM : Assigning pathogenicity to mitochondrial tRNA mutations: when ‘definitely maybe’ is not good enough. Trends Genet 2004; 20: 591–596.
Yarham JW, Al-Dosary M, Blakely EL et al: A comparative analysis approach to determining the pathogenicity of mitochondrial tRNA mutations. Hum Mutat 2011; 32: 1319–1325.
Lynn S, Wardell T, Johnson MA et al: Mitochondrial diabetes: investigation and identification of a novel mutation. Diabetes 1998; 47: 1800–1802.
Mansergh FC, Millington-Ward S, Kennan A et al: Retinitis pigmentosa and progressive sensorineural hearing loss caused by a C12258A mutation in the mitochondrial MTTS2 gene. Am J Hum Genet 1999; 64: 971–985.
Cardaioli E, Malfatti E, Da Pozzo P et al: Progressive mitochondrial myopathy, deafness, and sporadic seizures associated with a novel mutation in the mitochondrial tRNASer(AGY) gene. J Neurol Sci 2011; 303: 142–145.
Wong LJ, Yim D, Bai RK et al: A novel mutation in the mitochondrial tRNA(Ser(AGY)) gene associated with mitochondrial myopathy, encephalopathy, and complex I deficiency. J Med Genet 2006; 43: e46.
Leveque M, Marlin S, Jonard L et al: Whole mitochondrial genome screening in maternally inherited non-syndromic hearing impairment using a microarray resequencing mitochondrial DNA chip. Eur J Hum Genet 2007; 15: 1145–1155.
Lauber J, Marsac C, Kadenbach B, Seibel P : Mutations in mitochondrial tRNA genes: a frequent cause of neuromuscular diseases. Nucleic Acids Res 1991; 19: 1393–1397.
Wibom R, Hagenfeldt L, von Dobeln U : Measurement of ATP production and respiratory chain enzyme activities in mitochondria isolated from small muscle biopsy samples. Anal Biochem 2002; 311: 139–151.
Old SL, Johnson MA : Methods of microphotometric assay of succinate dehydrogenase and cytochrome c oxidase activities for use on human skeletal muscle. Histochem J 1989; 21: 545–555.
Hansson A, Hance N, Dufour E et al: A switch in metabolism precedes increased mitochondrial biogenesis in respiratory chain-deficient mouse hearts. Proc Natl Acad Sci USA 2004; 101: 3136–3141.
Hall RE, Henriksson KG, Lewis SF, Haller RG, Kennaway NG : Mitochondrial myopathy with succinate dehydrogenase and aconitase deficiency. Abnormalities of several iron-sulfur proteins. J Clin Invest 1993; 92: 2660–2666.
Stiggall DL, Galante YM, Hatefi Y : Preparation and properties of complex V. Methods Enzymol 1979; 55: 308–315, 819–821.
Taylor RW, Giordano C, Davidson MM et al: A homoplasmic mitochondrial transfer ribonucleic acid mutation as a cause of maternally inherited hypertrophic cardiomyopathy. J Am Coll Cardiol 2003; 41: 1786–1796.
Greaves LC, Yu-Wai-Man P, Blakely EL et al: Mitochondrial DNA defects and selective extraocular muscle involvement in CPEO. Invest Ophthalmol Vis Sci 2010; 51: 3340–3346.
Taylor RW, Taylor GA, Durham SE, Turnbull DM : The determination of complete human mitochondrial DNA sequences in single cells: implications for the study of somatic mitochondrial DNA point mutations. Nucleic Acids Res 2001; 29: E74–E74.
Tuppen HA, Hogan VE, He L et al: The p.M292T NDUFS2 mutation causes complex I-deficient Leigh syndrome in multiple families. Brain 2010; 133: 2952–2963.
Calvaruso MA, Smeitink J, Nijtmans L : Electrophoresis techniques to investigate defects in oxidative phosphorylation. Methods 2008; 46: 281–287.
Esteitie N, Hinttala R, Wibom R et al: Secondary metabolic effects in complex I deficiency. Ann Neurol 2005; 58: 544–552.
DiMauro S, Schon EA : Mitochondrial DNA mutations in human disease. Am J Med Genet 2001; 106: 18–26.
McFarland R, Taylor RW, Elson JL, Lightowlers RN, Turnbull DM, Howell N : Proving pathogenicity: when evolution is not enough. Am J Med Genet A 2004; 131: 107–108; author reply 109–110.
Levinger L, Oestreich I, Florentz C, Morl M : A pathogenesis-associated mutation in human mitochondrial tRNALeu(UUR) leads to reduced 3′-end processing and CCA addition. J Mol Biol 2004; 337: 535–544.
Sohm B, Frugier M, Brule H, Olszak K, Przykorska A, Florentz C : Towards understanding human mitochondrial leucine aminoacylation identity. J Mol Biol 2003; 328: 995–1010.
Bornstein B, Mas JA, Patrono C et al: Comparative analysis of the pathogenic mechanisms associated with the G8363A and A8296G mutations in the mitochondrial tRNA(Lys) gene. Biochem J 2005; 387: 773–778.
McFarland R, Schaefer AM, Gardner JL et al: Familial myopathy: new insights into the T14709C mitochondrial tRNA mutation. Ann Neurol 2004; 55: 478–484.
Tuppen HA, Fattori F, Carrozzo R et al: Further pitfalls in the diagnosis of mtDNA mutations: homoplasmic mt-tRNA mutations. J Med Genet 2008; 45: 55–61.
Perli E, Giordano C, Tuppen HA et al: Isoleucyl-tRNA synthetase levels modulate the penetrance of a homoplasmic m.4277T>C mitochondrial tRNA(Ile) mutation causing hypertrophic cardiomyopathy. Hum Mol Genet 2012; 21: 85–100.
Davidson MM, Walker WF, Hernandez-Rosa E, Nesti C : Evidence for nuclear modifier gene in mitochondrial cardiomyopathy. J Mol Cell Cardiol 2009; 46: 936–942.
Prezant TR, Agapian JV, Bohlman MC et al: Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet 1993; 4: 289–294.
Guan MX : Molecular pathogenetic mechanism of maternally inherited deafness. Ann NY Acad Sci 2004; 1011: 259–271.
Lu J, Qian Y, Li Z et al: Mitochondrial haplotypes may modulate the phenotypic manifestation of the deafness-associated 12S rRNA 1555A>G mutation. Mitochondrion 2010; 10: 69–81.
Kokotas H, Petersen MB, Willems PJ : Mitochondrial deafness. Clin Genet 2007; 71: 379–391.
Fischel-Ghodsian N, Kopke RD, Ge X : Mitochondrial dysfunction in hearing loss. Mitochondrion 2004; 4: 675–694.
Acknowledgements
We thank Drs Neil RM Buist, Robert D Steiner, Cary Harding and others who participated in the clinical evaluation, care and work-up of patient 2, and also provided valuable critique of this manuscript.
Funding: The project was supported by a Wellcome Trust Programme Grant (RWT) (Grant number 074454/Z/04/Z). The mitochondrial diagnostic laboratory in Newcastle is funded by the UK NHS Specialised Services to provide the ‘Rare Mitochondrial Disease of Adults and Children’ service (http://www.mitochondrialncg.nhs.uk/). Studies in Portland were supported in part by a grant from the Muscular Dystrophy Association (to NGK) and the Foundation Fighting Blindness (Columbia, MD) (to RGW). Financial support in Stockholm was given through the regional agreement on medical training and clinical research between Stockholm County Council and Karolinska Institutet.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Tuppen, H., Naess, K., Kennaway, N. et al. Mutations in the mitochondrial tRNASer(AGY) gene are associated with deafness, retinal degeneration, myopathy and epilepsy. Eur J Hum Genet 20, 897–904 (2012). https://doi.org/10.1038/ejhg.2012.44
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2012.44
Keywords
This article is cited by
-
Mitochondrial transcript maturation and its disorders
Journal of Inherited Metabolic Disease (2015)