Abstract
Osteoporosis is a metabolic bone disease with a strong genetic component. Family-based linkage studies were performed by a number of investigators to try to identify loci that might contain genes responsible for an increased susceptibility to osteoporosis. A whole-genome linkage scan using 400 microsatellite markers was performed in 27 members from two Maltese families with a highly penetrant form of osteoporosis. The phenotype was defined by lumbar and femoral z-scores calculated after measurement of bone mineral density by DEXA. Both males and females were among the affected individuals. Multipoint parametric and non-parametric linkage analyses were performed by EasyLinkage v4.01 using GENEHUNTER v2.1, assuming dominant and recessive modes of inheritance with variable penetrance. Evidence of linkage was observed to a marker at 11p12 where a non-parametric LOD score of 5.77 (P=0.0006) was obtained. A maximum heterogeneity LOD score of 2.55 for this region was obtained for the dominant mode of inheritance with 90% penetrance and a phenocopy rate of 1%. Following fine mapping, the critical interval was narrowed to a region that is 52.94 cM from 11p-telomere. In this region, the gene for tumour necrosis factor receptor-associated factor 6 (TRAF6) is located approximately 1 cM away from the indicated marker. Sequencing of the promoter region and exons of the TRAF6 gene revealed three sequence variants, one of which was found in three affected members within one family.
Similar content being viewed by others
Introduction
Osteoporosis (MIM 166710) is a polygenic disease where multiple gene variants, each having a small effect, contribute to the individual's increased susceptibility to the disease, although a major gene might also be involved.1 The search for genes responsible for complex diseases, such as osteoporosis, has presented a great challenge for geneticists during the last decade. Different approaches, such as case–control association and family-based linkage, have been used to identify any association between phenotype and gene variants.2
Family-based linkage studies were performed by a number of investigators to try to identify loci that might contain genes responsible for an increased susceptibility to osteoporosis. A number of chromosomal loci have been identified and confirmed to quantitative trait loci (QTL), such as bone mineral density (BMD), by whole-genome scans and by scanning candidate regions. Duncan et al3 observed suggestive linkage to the parathyroid hormone receptor type 1 (PTHR1) on chromosome 3p21 in a study carried out on a large group of British families. Moderate evidence of linkage was also reported at loci where other candidate genes are found including the COL1A1, COL2A1/VDR, IL-6 and oestrogen receptor 1. Wilson et al4 observed linkage with BMD at the lumbar spine (LS-BMD) with loci 3p21 and 1p36 as well as with other chromosomal loci including 11p, 2q and 19q, thus confirming the results obtained by Duncan et al3 and Devoto et al,5 where the latter associated locus 1p36 with low-hip BMD, after performing a non-parametric linkage analysis in seven extended families. Devoto et al6 confirmed these results after performing a higher resolution scan in an extended sample of 40 families coming from five populations of mixed ethnicity (Greeks, Italians, French, Jewish and Middle Eastern).
Another interesting locus is 11q12–13, which has been associated with peak bone mass7 as well as with other phenotypes including autosomal recessive osteopetrosis,8 high bone mass phenotype9 and osteoporosis pseudoglioma syndrome.10 A number of loci were confirmed by linkage to a QTL, such as BMD at different anatomical sites,11, 12 variation in femoral structure13 and peak bone mass reached early in life.14
Linkage and association were confirmed to chromosome 20p12 in a study carried out on the Icelandic population.15 Discrete phenotypes were defined by using z-scores corrected for age and combined with fractures of different degrees of severity. Linkage was observed at chromosome 20p12. Linkage disequilibrium mapping using a number of single-nucleotide polymorphisms (SNPs) at this region indicated a missense polymorphism and two SNP haplotypes within the bone morphogenic protein-2 (BMP2) gene, which were associated with osteoporosis in both the Icelandic and Danish populations.15 This region was later confirmed in a study of European Caucasians where evidence of linkage to regions 11q23 and Xq27 was also observed.16
In this study, a whole-genome-wide scan was performed in two extended Maltese families with a highly penetrant form of osteoporosis, for possible identification of loci that might be responsible for osteoporosis in the Maltese population.
Materials and methods
Subjects
Two extended families with a high incidence of osteoporosis were included in this study (Figures 1 and 2). These families were recruited from the Department of Obstetrics and Gynaecology, St Luke's Hospital, Malta and selected from probands that already participated in previous association studies.17, 18 These two probands were observed to be severely osteoporotic according to WHO criteria (t-score<−2.50), where both probands had t-scores of −3.50, were relatively young in age when diagnosed (50 and 55 years, respectively) and have a known family history of osteoporosis. A total of 27 family members were recruited, nine of whom came from pedigree 1 and 18 from pedigree 2 (Table 1). All participants were asked to sign an informed consent form and answered a detailed questionnaire concerning medical history, medication, lifestyle and dietary habits as well as detailed information about family history of bone disease, osteoporosis and fracture. This study was approved by the Research Ethics Committee of the Faculty of Medicine and Surgery, University of Malta.
Pedigree 1 showing inheritance of part of chromosome 11. Affected individuals (black shaded) in generation II where defined according to the WHO criteria (t-score <−2.5). In generation III, affected status was defined by z-score <−1.0. Individuals II:3 and II:4 were not recruited, therefore affected status is unknown. DNA was not obtained from individual III:5. Inferred genotypes were generated by GENEHUNTER and are shown in parentheses.
This figure shows pedigree 2 and inheritance of part of chromosome 11. Affected individuals (black shaded) in generation II where defined according to the WHO criteria (t-score <−2.5). In generation III, affected status was defined by z-score <−1.0. Individuals II:3, II:7, III:3, III:12 and III:15 were not recruited, therefore affected status is unknown. Inferred genotypes were generated by GENEHUNTER and are shown in parentheses.
BMD at the LS and femoral neck (FN) of all participants was measured by DEXA using a Norland 385 (Norland, Medical Systems Inc.) at the Bone Density Unit, St Luke's Hospital, Malta. Peripheral blood was withdrawn from all participants for DNA analysis and biochemical tests in order to exclude secondary osteoporosis. Biochemical tests performed included serum creatinine, urea, random blood glucose, calcium (total and ionised), magnesium, phosphate, total cholesterol, total bilirubin, alkaline phosphatase, γ-glutamyl transferase, alanine transaminase together with a complete blood count. All biochemical tests were performed using a Hitachi 917 analyser (Boehringer Mannheim, Germany) and a Sysmex K4500 haematology analyser was used for the complete blood count. None of the family members were observed to have medical conditions or were on medications known to affect bone metabolism.
Phenotype definition
Five phenotype scenarios were initially analysed in this study. In scenario I, affected individuals for a low BMD phenotype (for chronological age) were defined by age- and sex-adjusted z-scores at LS and/or FN of less than −1.0. A more severe phenotype was defined by z-scores of less than −1.5, in scenario II. These z-scores were calculated by using reference data of BMD measurements obtained for sex- and age-matched normal individuals of the same ethnicity (Caucasian).
The phenotype was also defined using the guidelines suggested by the International Society of Clinical Densitometry,19 where osteoporosis for postmenopausal women and men over 50 years of age was defined using a lumbar and/or femoral t-score of less than −2.5 (as recommended by the WHO). On the other hand, z-scores were used for younger individuals where a z-score of less than −1.0 was used as the threshold for an affected status and that of less than −2.0 for a more severe phenotype, for scenarios III and IV, respectively.
For scenario V, analysis was performed, by assuming as affected only those individuals having femoral z-scores of less than −1.0.
The same five scenarios were further analysed by assuming that all unaffected individuals had an unknown phenotype. This assumption takes into consideration the possibility that any apparently clinically unaffected individuals might be actually affected, thus reducing the chance of obtaining false negative results.
DNA analysis
DNA was extracted and purified from peripheral blood leucocytes by a salting out procedure, adapted from Miller et al.20 Four hundred short tandem repeats (STRs) spread across the 22 autosomes and x-chromosome with an average spacing of 8.63 cM and heterozygosity of 0.77 were selected for the initial scan. Genotyping of microsatellite markers was performed by polymerase chain reaction (PCR), and fragment analysis of amplified products was performed using a 3730xl ABI genetic analyzer (PE Applied Biosystems, Foster City, CA). The average performance of the markers for all samples was of 96.96%. Genotyping was performed commercially at the McGill University and Genome Quebec Innovation Centre, Quebec, Canada.
Linkage analysis
Multipoint parametric and non-parametric linkage analyses were performed by EasyLinkage v4.01 (http://www.uni-wuerzburg.de/nephrologie/molecular_genetics/molecular_genetics.htm),21 using the software GENEHUNTER v2.1 and GENEHUNTER v1.2 to calculate Zlr scores using Kong and Cox model.22 The Zlr algorithm addresses the problem with previous versions of GENEHUNTER where non-parametric LOD (NPL) scores are found to be too conservative when inheritance data are not complete.
Parametric analysis was carried out using variable penetrances for both a dominant and recessive mode of inheritance. Penetrances used for the dominant model were 0.01, 0.90 and 0.90 for the wild-type homozygote, mutant heterozygote and mutant homozygote, respectively. The recessive model was defined by penetrances 0.01, 0.01 and 0.80 for the wild-type homozygote, mutant heterozygote and mutant homozygote, respectively. A more complex model was also analysed using penetrances 0.01, 0.05, 0.30 for wild-type homozygotes, mutant heterozygotes and mutant homozygotes, respectively. For all models, the disease allele prevalence frequency assumed was of 0.001, and phenocopy rate of 1%. A parametric analysis assuming heterogeneity was computed using data from both families heterogeneity LOD (HLOD). A codominant allele frequency algorithm was used for the analysis, as suggested by the EasyLinkage user manual, for extended families. Following the initial scans, loci observed to have a high NPL and LOD score were further analysed using variable penetrances and phenocopy rates. These analyses were carried out for all the phenotypes described above, using the sex-averaged deCode genetic map.
Analysis of candidate gene
The eight exons of the tumour necrosis factor receptor-associated factor 6 (TRAF6) (MIM 602355) gene, including exon–intron boundaries and 1 kb upstream from the transcriptional start site, were amplified by PCR in two severely affected individuals and two normal individuals. Forward and reverse primers were designed using the reference sequence of the transcript ENST00000313105 (http://www.ensembl.org). Sequencing reactions were performed using the Big Dye Terminator Kit v3.1 and ABI 3130 genetic analyzer (PE Applied Biosystems, Foster City, CA), at MLS BioDNA Ltd, Malta.
Sequence variants identified were analysed in all family members and a cohort of unrelated postmenopausal women,17, 18 either by direct sequencing or by restriction fragment length polymorphism (RFLP). PCR products (10 μl) were digested using appropriate restriction enzymes according to the manufacturer's instructions (New England Biolabs, Beverly, MA, USA). Agarose gel electrophoresis was carried out using 15 μl of digested product. Following electrophoresis and staining with ethidium bromide, digested products were viewed and photographed under ultraviolet illumination (Polaroid DS-34). Nomenclature used refers to the actual nucleotide changes in the respective polymorphisms.
Results
The clinical and general characteristics of the individuals from both families are shown in Tables 1 and 2. A low BMD for chronological age phenotype was defined according to WHO criteria for an age- and sex-adjusted z-score of less than −1.0 at the LS and/or FN. Pedigree 1 is shown in Figure 1, with individual II:1 as the proband, whose recruited siblings were all found to be affected, with the most severe individual (II:8) having lumbar t-score and z-score of −5.68 and −3.09, respectively. All daughters of the proband had very low BMD. Her 35-year-old daughter (III:2) was found to have lumbar t-score and z-score of −2.49 and −2.29, respectively. Pedigree 2 (Figure 2) was identified from a 55-year-old woman (II:1), who was osteoporotic at the LS (t-score −3.47). Her youngest sister (II:6) was found to be severely osteoporotic at both the lumbar and FN with t-scores of −3.75 and −2.72, respectively. Her other two sisters (II:4 and II:5) were also found to be severely osteoporotic but had z-scores >−1.0. Low BMD was observed to occur even in males within this family. The 29-year-old son of the proband (III:1) had a lumbar t-score of −2.00 and z-score of −2.02. His 37-year-old cousin (III:10) was also severely affected for his age having a lumbar t-score of −2.32 and z-score of −2.19.
From the initial genome-wide scan in both families, evidence of linkage was observed to marker D11S1392 where a NPL score of 5.77 (P=0.0006) was obtained. A maximum HLOD score of 2.55 for this region was reached for the dominant mode of inheritance with 90% penetrance and phenocopy rate of 1%.
The same marker showed evidence of linkage when analysing families according to their femoral status (scenario V). For this phenotype, an NPL of 5.24 (P=0.0020) and an HLOD of 2.12 was obtained for this marker assuming a dominant mode of inheritance with 70% penetrance. This was the only locus that showed evidence of linkage to this phenotype with an affected status defined by femoral z-scores only.
Marker D5S1960 also showed evidence of linkage when a recessive mode of inheritance with 80% penetrance was analysed. Highest HLOD for this model was of 2.78 with a NPL of 5.10 (P=0.0031).
High-resolution scan
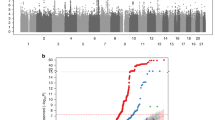
A high-resolution scan was performed by adding four additional markers at 1–2 cM intervals at the indicated locus on chromosome 11 (D11S4101, D11S935, D11S4102 and D11S1911). Linkage was confirmed to this region on chromosome 11, where an HLOD of 3.07, NPL of 7.0 (P=0.0014) and Zlr score of 3.01, were obtained to marker D11S935 that is 52.94 cM from 11p-telomere (in scenario 1). Figure 3 shows the LOD plot following fine mapping where the critical interval was narrowed after the addition of markers in this region. Table 3 shows the contribution of each individual family whereas the results for both families are shown in Table 4. Consistent results were obtained at this region on chromosome 11p12 with a highest Zlr score of 3.74 in scenarios IV and IX.
This locus was further analysed by varying the penetrance and phenocopy rates for the dominant mode of inheritance. Results obtained are shown in Table 5, where it can be observed that the highest HLOD score was of 3.32 at penetrances of 0.8 and 0.7 with a phenocopy rate of 5%.
When performing the same analysis on pedigrees assuming clinically unaffected individuals as having an unknown phenotype (scenarios VI–X), highest HLOD and NPL scores obtained were of 3.35 and 6.9 (P=0.0002), respectively, to marker D11S935 for the dominant mode of inheritance with 90% penetrance and phenocopy 1% (scenario IX).
TRAF6 sequencing
Candidate genes found within the linkage interval were selected with prior knowledge of bone physiology using the NCBI map viewer (http://www.ncbi.nlm.nih.gov) and freely available web tool The GeneSeeker v2.0 (http://www.cmbi.kun.nl/GeneSeeker/). The whole area from 49 to 55 cM on chromosome 11 (deCode genetic map) was searched for possible genes that might be involved in the disease. A total of 22 genes and two hypothetical proteins are known to be found in this region, with the best candidate being the gene coding for the TRAF6 (MIM 602355), that is approximately 1 cM away from marker D11S935.
Following direct sequencing, three different variants were identified when compared to reference sequences on the NCBI and Ensembl databases. An A–T transversion was identified at position −721 (5′ upstream of exon 1), when compared to TRAF6 reference sequence (AY228337) (Figure 4). This variant was not described previously. Following sequencing of all family members, three affected individuals from pedigree 1 were observed to be heterozygous for this variant. Individuals from pedigree 2 were all wild-type homozygotes. RFLP was carried out in 82 unrelated postmenopausal women, using restriction enzyme HindIII, which was able to cut the PCR product in the presence of the T allele. This variant was observed to be very rare within an unrelated group of postmenopausal women, as only three heterozygotes were observed. After screening 350 chromosomes in a random sample from the general population, only two alleles were observed (0.57%) with this variant having a population frequency of 1.1%.
An already known insertion/deletion of a T in the intron between exons 4 and 5, in the polyT region 16 bp ahead of the exon–intron boundary (rs3830511), was identified. When analysing all family members and controls, only three individuals were observed to be heterozygotes for this insertion/deletion, one of whom was severely affected and the other two were normal individuals.
A transition from G to A was found in the intron between exons 6 and 7, 110 bp upstream of the exon–intron boundary. When sequencing all members from both families, four heterozygotes for this variant were identified, the rest were homozygous for the wild-type allele G. Three of the four heterozygous individuals for this variant were affected individuals, two of whom were also heterozygous for the T insertion/deletion described above. Genotyping by RFLP (PvuII) was performed in 82 unrelated postmenopausal women. Genotype frequencies observed were 72.3% GG, 26.5% GA and 1.2% AA.
Discussion
In this study, a novel locus on chromosome 11 was linked with osteoporosis in two extended Maltese families. Following fine mapping, the critical region was reduced to a 4 cM region at 11p12 between markers D11S1392 and D11S4102. As shown in Figures 1 and 2, inherited haplotypes at this locus can be observed in both individual families. This seems to be inherited in an autosomal dominant fashion in affected individuals, with a number of recombination events occurring very close to this region. In pedigree 2 (Figure 2), an affected male (III:4) does not carry this causative haplotype suggesting that low BMD in this individual might be due to an environmental factor (phenocopy). On the other hand, the suspected haplotype is present in two apparently clinically normal individuals (III:2 and III:7). This could be due to the fact that individual III:2 is only 23 years old and perhaps is still too young to be affected. Individual III:7 has a high BMI that might be modifying the effect of the causative gene. Also, other genetic and/or environmental factors might be modifying the effect of the causative gene in this region leading to incomplete penetrance of the disease allele. By assuming a reduced penetrance and increased phenocopy rate, more significant results were obtained at this locus (Table 5).
From this study, it is evident that multiple genes are involved in the pathogenesis of osteoporosis in these two families. Besides the most significant region on chromosome 11p12, another region on chromosome 5 gave an NPL of 5.10 and HLOD of 2.78 to marker D5S1960. Other interesting regions that require further investigation were found on chromosomes 3, 6, 9 and 17. Linkage to chromosomes 3q and 17q22 was also reported by Koller et al,13 where strong evidence of linkage to chromosome 17q22 was reported with variations in femoral structure. Among the candidate genes involved in bone physiology on chromosome 17q22 are those coding for type I collagen (COL1A1) and noggin.23
No evidence of linkage was observed to previously indicated regions such as those on chromosomes 11q12–13, 1p36,5, 6, 7, 8, 9, 10 20p12,15 11p23 and Xq27.16 Weak evidence of linkage with whole body BMD was observed by Wilson et al4 to a region that is 10 cM away from our indicated region on chromosome 11p12, which is approximately 20 cM away from 11q12–13 indicated in previous studies which is well known to contain genes responsible for other bone phenotypes,9, 10 such as those coding for the low-density lipoprotein receptor-related protein 5 and the T-cell immune regulator 1.
In a multifactorial disease, such as osteoporosis, lack of concordance between different studies might be due to a multitude of environmental factors including dietary and lifestyle factors, possibly modifying the effect of the causative gene. Apart from this, linkage analysis is affected by incomplete penetrance, phenocopy and genetic heterogeneity, where different genes might be responsible for disease in different ethnic groups.
Differences in study design, approach and analysis of data that might result in type I errors, are other reasons for lack of replication of results. Our study is the second published study using a qualitative instead of a quantitative type of analysis for osteoporosis. This approach has also been used by Styrkarsdottir et al15 in a large study performed in the Icelandic population, where variants within the BMP-2 gene on chromosome 20p12 were associated with the disease. In previous studies, linkage was most often sought by using BMD measurements at different anatomical site as a QTL.3, 4, 5, 6, 7, 11, 12, 13, 14, 16 This approach is not always advantageous especially as it is known that BMD is not the only determinant of osteoporosis and fracture risk. Each individual trait, such as BMD and other radiographic measurements is more likely to be affected by a number of loci, each having a very small effect on the phenotype. This makes it very difficult to detect linkage unless a larger sample size is used. However, when different genome scans for a number of complex diseases were compared for evaluation, it was shown that highly significant linkage was obtained from studies using a qualitative approach and significant results were not always achieved by the largest studies.24
Our study was carried out using only two large pedigrees in which osteoporosis appeared to exhibit classical Mendelian inheritance. Other loci were linked to other human diseases in studies using single extended families with multiple affected individuals such as those for an autosomal dominant keratoconus25 and for the localisation of a cholesterol-lowering gene.26 An appropriate study design is very important for the successful localisation of genes responsible for a genetically heterogeneous disease such as osteoporosis. For heterogeneous disease, power is increased by using large extended families especially those coming from an isolated population rather than from an outbred one.27 Two examples of isolated populations are the Icelandic and Finnish, where in the latter a number of genes and chromosomal loci were confirmed.28 The BMP2 gene on chromosome 20p12 was identified in a study carried out in the isolated population of Iceland.15
The observation that common haplotypes were found in all affected members of individual families together with the high HLOD of 3.32 and NPL score of 7.0, points to a novel locus for osteoporosis on 11p12. This locus has been also linked to a number of inflammatory disorders. Amos et al29 showed evidence of linkage to 11p12 with rheumatoid arthritis in 642 Caucasian families. Apart from rheumatoid arthritis, this locus was also linked with coeliac disease30 and inflammatory bowel disease.31Such observations might support the hypothesis that a common inflammatory process controlled by genes found at this locus might be responsible for the onset of these diseases. This indicates that osteoporosis might also be caused by a mild subclinical inflammation that is triggered with advancing age.32
TRAF6 plays a very important role in the differentiation and activation of osteoclasts when it interacts with receptor activator of NF-κβ (RANK) and activates transcription mediated by activator protein-1.33, 34 The importance of TRAF6 in osteoclast function was shown from experiments on knockout mice where osteopetrosis was observed in TRAF6-deficient mice.35 The RANKL/RANK system and therefore TRAF6 plays a very important role in what is now known as osteoimmunology. Takayanagi36 describes how immunology and bone physiology interact and share various molecules. TRAF6 together with other TRAFs are recruited by RANK upon stimulation by RANKL, which is also expressed by T cells, and controlled by signal transducers and activators of transcription through signalling from the interferon-γ receptor. From our DNA sequencing analysis, we identified three sequence variants in the TRAF6 gene, one in the promoter region and the other two in intervening introns. The variant found in the promoter region was detected in three affected individuals in one family and was observed to be very rare in the population. Further investigation of this variant is being conducted to address its possible role on gene expression.
No variants were found in the coding regions of TRAF6, showing that its sequence is conserved, thus indicating its important biological role. Recently, Lappalainen et al37 identified an insertion/deletion in a polyT stretch in intron 3, a similar polymorphism to the one we identified in intron 4, but failed to find association with inflammatory bowel disease. As these polymorphisms are found in the polyT region 3′ upstream from the intron–exon boundary, one cannot exclude their possible role in the splicing mechanism. Sequence variants identified in this study do not seem to be completely linked with the disease in these two families. This does not completely exclude its possible role in the pathogenesis of osteoporosis, as other variants might be present in regions that were not sequenced (introns). Also different genes might be responsible for the disease in each individual family. This might be suggested by highest LOD scores for pedigree 1 being closer to the TRAF6 gene (∼54 cM), whereas those observed in pedigree 2 were approximately 2 cM upstream (∼52 cM) (Table 3). Also different alleles were observed in the two inherited haplotypes in the two families. Another possible candidate gene found closer to the 52 cM region is the gene coding for CD44, which was also observed to play a role in inflammatory bone loss.38
These results suggest a possible role of a gene on chromosome 11p12 that might be responsible for osteoporosis in these families. Sequence variants within the TRAF6 gene and other genes found at this locus might be responsible for this type of osteoporosis. Further studies of this region will help us to understand better the pathophysiology of osteoporosis and may lead to the development of more effective treatments.
References
Cardon LR, Garner C, Bennett ST et al: Evidence for a major gene for bone mineral density in idiopathic osteoporotic families. J Bone Miner Res 2000; 15: 1132–1137.
Huang QY, Recker RR, Deng HW : Searching for osteoporosis genes in the post-genome era, progress and challenges. Osteoporos Int 2003; 14: 701–715.
Duncan EL, Brown MA, Sinsheimer J et al: Suggestive linkage of the parathyroid receptor type 1 to osteoporosis. J Bone Miner Res 1999; 14: 1993–1999.
Wilson SG, Reed PW, Bansal A et al: Comparison of genome screens for two independent cohorts provides replication of suggestive linkage of bone mineral density to 3p21 and 1p36. Am J Hum Genet 2003; 72: 144–155.
Devoto M, Shimoya K, Caminis J et al: First-stage autosomal genome screen in extended pedigrees suggests genes predisposing to low bone mineral density on chromosomes 1p, 2p and 4q. Eur J Hum Genet 1998; 6: 151–157.
Devoto M, Specchia C, Li HH et al: Variance component linkage analysis indicates a QTL for femoral neck bone mineral density on chromosome 1p36. Hum Mol Genet 2001; 10: 2447–2452.
Koller DL, Rodriguez LA, Christian JC et al: Linkage of a QTL contributing to normal variation in bone mineral density to chromosome 11q12-13. J Bone Miner Res 1998; 13: 1903–1908.
Heaney C, Shalev H, Elbedour K et al: Human autosomal recessive osteopetrosis maps to 11q13, a position predicted by comparative mapping of murine osteosclerosis (oc) mutation. Hum Mol Genet 1998; 7: 1407–1410.
Johnson ML, Gong G, Kimberling W, Recker SM, Kimmel DB, Recker RR : Linkage of a gene causing high bone mass to human chromosome 11 (11q12–13). Am J Hum Genet 1997; 60: 1326–1332.
Gong Y, Vikkula M, Boon L et al: Osteoporosis-pseudoglioma syndrome, a disorder affecting skeletal strength and vision, is assigned to chromosome region 11q12-13. Am J Hum Genet 1996; 59: 146–151.
Kammerer CM, Schneider JL, Cole, SA et al: Quantitative trait loci on chromosomes 2p, 4p, and 13q influence bone mineral density of the forearm and hip in Mexican Americans. J Bone Miner Res 2003; 18: 2245–2252.
Wynne F, Drummond FJ, Daly M et al: Suggestive linkage of 2p22-25 and 11q12-13 with low bone mineral density at the lumbar spine in the Irish population. Calcif Tissue Int 2003; 72: 651–658.
Koller DL, Liu G, Econs MJ et al: Genome screen for quantitative trait loci underlying normal variation in femoral structure. J Bone Miner Res 2001; 16: 985–991.
Econs MJ, Koller DL, Hui SL et al: Confirmation of linkage to chromosome 1q for peak vertebral bone mineral density in premenopausal white women. Am J Hum Genet 2004; 74: 223–228.
Styrkarsdottir U, Cazier JB, Kong A et al: Linkage of osteoporosis to chromosome 20p12 and association to BMP2. PLoS Biol 2003; 1: 1–10.
Shen H, Zhang YY, Long JR et al: A genome-wide linkage scan for bone mineral density in an extended sample: evidence of linkage on 11q23 and Xq27. J Med Genet 2004; 41: 743–751.
Vidal C, Grima C, Brincat M, Megally N, Xuereb-Anastasi A : Associations of polymorphisms in the vitamin D receptor gene (BsmI and FokI) with bone mineral density in postmenopausal women in Malta. Osteoporos Int 2003; 14: 923–928.
Vidal C, Brincat M, Xuereb-Anastasi A : TNFRSF11B gene variants and bone mineral density in postmenopausal women in Malta. Maturitas 2005; 53: 386–395.
Khan AA, Bachrach L, Brown JP et al: Standards and guidelines for performing central dual-energy X-ray absorptiometry in premenopausal women, men, and children. J Clin Densitom 2004; 7: 51–64.
Miller SA, Dykes DD, Polesky HF : A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215.
Lindner TH, Hoffmann K : EasyLinkage: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics 2005; 21: 405–407.
Kong A, Cox NJ : Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 1997; 61: 1179–1188.
Wu XB, Li Y, Schneider A et al: Impaired osteoblastic differentiation, reduced bone formation, and severe osteoporosis in noggin overexpressing mice. J Clin Invest 2003; 112: 924–934.
Atmuller J, Palmer LJ, Fischer G, Scherb H, Wjst M : Genomewide scans of complex human diseases: true linkage is hard to find. Am J Hum Genet 2001; 69: 836–950.
Brancati F, Valente EM, Sarkozy A et al: A locus for autosomal dominant keratoconus maps to human chromosome 3p14-q13. J Med Genet 2004; 41: 188–192.
Knoblauch H, Muller-Myhsok B, Busjahn A et al: A cholesterol lowering gene maps to chromosome 13q. Am J Hum Genet 2000; 66: 157–166.
Sillanpaa MJ, Auranen K : Replication in genetic studies of complex traits. Ann Hum Genet 2004; 68: 646–657.
Peltonen L : Positional cloning of disease genes: advantages of genetic isolates. Hum Hered 2000; 50: 66–75.
Amos CI, Chen WV, Lee A et al: High density SNP analysis of 642 Caucasian families with rheumatoid arthritis identifies two new linkage regions on 11p12 and 2q33. Genes Immun 2006; 7: 277–286.
King AL, Fraser JS, Moodie SJ et al: Celiac disease: follow-up linkage study provides further support for existence of a susceptibility locus on chromosome 11p11. Ann Hum Genet 2001; 65: 377–386.
Paavola-Sakki P, Ollikainen V, Helio T et al: Genome-wide search in Finnish families with inflammatory bowel disease provides evidence for novel susceptibility loci. Eur J Hum Genet 2002; 11: 112–120.
Ginaldi L, Di Benedetto MC, De Martinis M : Osteoporosis, inflammation and ageing. Immun Ageing 2005; 2: 14.
Teitelbaum SL : RANKing c-Jun in osteoclast development. J Clin Invest 2004; 114: 463–465.
Armstrong AP, Tometsko ME, Glaccum M et al: A RANK/TRAF6-dependent signal transduction pathway is essential for osteoclast cytoskeletal organization and resorptive function. J Biol Chem 2002; 277: 44347–44356.
Lomaga MA, Yeh WC, Sarosi I et al: TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signalling. Genes Dev 1999; 13: 1015–1024.
Takayanagi H : Mechanistic insight in osteoclast differentiation in osteoimmunology. J Mol Med 2005; 83: 170–179.
Lappalainen M, Paavola-Sakki P, Halme L et al: Screening of tumor necrosis factor receptor-associated factor 6 as candidate gene for inflammatory bowel disease. Scand J Gastroenterol 2006; 41: 424–429.
Hayer S, Steiner G, Gortz B et al: CD44 is a determinant of inflammatory bone loss. J Exp Med 2005; 201: 903–914.
Acknowledgements
First of all we thank the families that participated in this study for their collaboration. We also thank Dr Andrei Verner and the staff of the Genotyping Facility McGill University and Genome Quebec Innovation Centre, Montreal, Quebec, Canada for the STR genotyping. We thank the Pathology Department, St Luke's Hospital, G’Mangia for supporting us to carry out the biochemical and haematology tests. This work was supported by the Research Fund Committee, University of Malta.
Author information
Authors and Affiliations
Corresponding author
Additional information
Web Resources and Software
Online Mendelian Inheritance in Man (OMIM) http://www.ncbi.nlm.nih.gov/Omim/ (search for osteoporosis, TRAF6)
Ensembl Gene Database (http://www.ensembl.org)
GeneSeeker v2.0 (http://www.cmbi.kun.nl/GeneSeeker/) searching for genes expressed in bones, osteoblasts, osteoclasts and skeleton
The software Cyrillic v2.1.3 (Cherwell Scientific Publishing Ltd) was used to design pedigrees in Figures 1 and 2.
Rights and permissions
About this article
Cite this article
Vidal, C., Galea, R., Brincat, M. et al. Linkage to chromosome 11p12 in two Maltese families with a highly penetrant form of osteoporosis. Eur J Hum Genet 15, 800–809 (2007). https://doi.org/10.1038/sj.ejhg.5201814
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201814
Keywords
This article is cited by
-
Genetic association study identified a 20 kb regulatory element in WLS associated with osteoporosis and bone mineral density in Han Chinese
Scientific Reports (2017)
-
A genome-wide association analysis implicates SOX6 as a candidate gene for wrist bone mass
Science China Life Sciences (2010)
-
Osteoporosis as an Hereditary Disease
Clinical Reviews in Bone and Mineral Metabolism (2010)