Abstract
Mutations in PQBP1 were recently identified in families with syndromic and non-syndromic X-linked mental retardation (XLMR). Clinical features frequently associated with MR were microcephaly and/or short stature. The predominant mutations detected so far affect a stretch of six AG dinucleotides in the polar-amino-acid-rich domain (PRD), causing frameshifts in the fourth coding exon. We searched for PQBP1 exon 4 frameshifts in 57 mentally retarded males in whom initial referral description indicated at least one of the following criteria: microcephaly, short stature, spastic paraplegia or family history compatible with XLMR, and in 772 mentally retarded males not selected for specific clinical features or family history. We identified a novel frameshift mutation (23 bp deletion) in two half-brothers with specific clinical features, and performed prenatal diagnosis in this family. We also found two different 21 bp in-frame deletions (c.334–354del(21 bp) and c.393–413del(21 bp)) in four unrelated probands from various ethnic origins, each deleting one of five copies of an imperfect seven amino-acid repeat. Although such deletions have not been detected in 1180 X chromosomes from European controls, the c. 334–354del(21 bp) was subsequently found in two of 477 Xs from Indian controls. We conclude that pathogenic frameshift mutations in PQBP1 are rare in mentally retarded patients lacking specific associated signs and that the 21 bp in-frame deletions may be non-pathogenic, or alternatively could act subtly on PQBP1 function. This touches upon a common dilemma in XLMR, that is, how to distinguish between mutations and variants that may be non-pathogenic or represent risk factors for cognitive impairment.
Similar content being viewed by others
Introduction
X-linked mental retardation (XLMR) has been proposed to account for the observed 30% excess of males in schools or institutions for mentally handicapped. 1 Intense research in recent years has led to the identification of many genes mutated in syndromic or non-syndromic forms of XLMR. However, for several of these genes it has been shown that they can be found mutated in both clearly syndromic or apparently non-syndromic families or individual patients, suggesting a ‘vanishing boundary’ between these two categories.2, 3 This has important implications for diagnostic purposes: while definition of a syndrome associated with mutations in a given gene can lead to a clinical selection of patients (even sporadic cases) in whom such mutations can be sought with good efficiency (as for instance for the Coffin–Lowry or ATRX syndromes), in the case of ‘non-syndromic’ mental retardation (MR), one can at present only search for mutations in XLMR genes in families with clear X-linked segregation, as the probability of finding a mutation in any given XLMR gene in a sporadic case is very low.3, 4 It is thus important to know the range of phenotypes associated with mutations in an XLMR gene, and the incidence of such mutations in patients pre-selected or not selected for specific features.
Mutations in the PQBP1 gene coding for the polyglutamine-binding protein 1 were initially identified in seven families with XLMR,5, 6 and subsequently in four additional families.7, 8 The clinical features most frequently associated with mental retardation (MR) are microcephaly (86% of patients) and/or short stature (58%).7, 9 Other signs, such as spasticity, small testes, anal atresia or stenosis, cardiac defects, ocular coloboma, cleft palate and dysmorphic facial features are observed in a smaller proportion of patients.5, 7, 9 One of these families was the original family defining the Renpenning syndrome, characterized by MR associated with microcephaly, short stature and small testes,6, 10 one corresponds to the Sutherland and Haan syndrome family,6, 11 and another one to the Golabi–Ito–Hall syndrome, characterized by MR associated with microcephaly, short stature and ectodermal manifestations.7 Considerable inter- and intrafamilial phenotypic variation was reported, which ranged from severe MR, microcephaly, spasticity, short stature, craniofacial abnormalities and congenital heart defect corresponding to the Hamel cerebropalatocardiac syndrome in one family (N40),12 to a milder nonspecific form in another family (MRX55),5 in whom all three affected males presented with moderate MR and were initially reported to be without any specific features, except for short stature in one patient.13 Heterozygous females usually do not present clinical signs, but some of them show skewed X-inactivation.7, 8
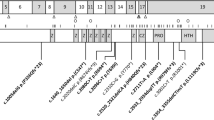
The PQBP1 gene comprises six exons and encodes a protein of 265 amino acids. Three alternative transcripts, that would correspond to shorter proteins, have been described.14 The PQBP1 protein was identified in a yeast two-hybrid screen using the homopolymeric glutamine repeats (Poly Q) of mouse transcription factor Brn-2 (POU3F2) as a bait.15 In vitro and in vivo studies have shown that PQBP1 binds to huntingtin and ataxin 1 through their polyglutamine repeats.16, 17 The full-length PQBP1 protein is localized predominantly in the nucleus15, 18, 19 and is characterized by several domains: an acidic region, a WW domain (a domain characterized by two conserved tryptophans) that may regulate gene expression,17, 20, 21, 22 a polar amino-acid-rich-region domain (PRD) (containing a DR/ER stretch) that is essential for polyglutamine binding and may be involved in transcriptional control by interfering with Brn2,15, 16 a nuclear localization signal (NLS), and a C2-domain that has been shown to bind to a component of the nuclear pre-mRNA splicing machinery.23, 24 The PRD and the NLS domains are both encoded by exon 4.14
The predominant mutations detected so far (8/11 families) affect a stretch of six AG dinucleotides in the DR/ER repeat in the PRD, and these include insertion of an AG dinucleotide, deletion of a single AG unit or deletion of two AG dinucleotides, all causing frameshifts in exon 4.5, 7, 8 These mutations result in proteins lacking a part of the DR/ER repeat in the PRD, the entire NLS and the C2 domain. Cell-transfection experiments showed that the corresponding mutant PQBP1 proteins were dispersed throughout the cell and may no longer interact with Brn-2.5 A further study revealed another AG dinucleotide deletion at the end of exon 4 (c.575–576 del AG) and a one bp insertion in exon 5 (c.641insC).6 More recently, a missense mutation has been identified in the WW domain (p.Y65C) in the Golabi–Ito–Hall syndrome family.7
As nine of the eleven mutations identified so far lead to an easily detectable size change in exon 4 of the PQBP1 gene, we wished to test the implication of such mutations in a cohort of 772 mentally retarded males who have not been selected for specific clinical features, and in 57 patients in whom initial referral description indicated that they had at least one of the following criteria: microcephaly, short stature, spastic paraplegia, or a family history compatible with XLMR. We report here a novel frameshift mutation (a 23 bp deletion), found in two half-brothers presenting with specific clinical features already described in several patients with PQBP1 mutations, and two different in-frame 21 bp deletions (each deleting one of five copies of an imperfect seven amino-acid repeat in the PRD domain), found in four unrelated patients. As we also found one of these 21 bp deletions in some ethnically matched controls, we discuss their pathogenic significance.
Patients and methods
Patients
We screened 772 males (254 of Indian origin) with MR, who had not been selected for the presence of other clinical features, and who were initially referred for systematic Fragile X syndrome testing. They had various forms and severities of MR but associated clinical data were not always available. We also screened a cohort of 57 selected males with MR also initially referred for Fragile X testing, but in whom available clinical description indicated a family history compatible with XLMR transmission (n=17) or association with at least one of the following criteria: microcephaly, short stature, or spastic diplegia (n=40). All 829 patients were negative for Fragile X syndrome or major aneuploidies.
We also tested 1265 control X chromosomes, 680 from European individuals, 85 from Pakistani individuals, 23 from Turkish individuals and 477 from Indian individuals, all of normal intelligence.
Methods
DNA extraction from blood samples was performed by standard procedures. PCR amplifications of PQBP1 exon 4 were performed using primers reported earlier.5 PCR reactions were carried out in 25 μl reaction volumes containing 100 ng of genomic DNA, 20 pmol of each primer, and 1 U Taq Sigma (Roche). PCR amplification was performed in a Whatman Biometra/MJ Research thermocycler with the following conditions: 95°C for 5 min, 25 cycles of 95°C for 30 s, 59°C for 30 s, and 72°C for 1 min, and a final extension step of 72°C for 10 min. Fluorescent PCR products were analyzed by fragment size determination on an ABI310 or an ABI3100 automated capillary sequencer (Applied Biosystems, Foster city, CA, USA). Non-fluorescent PCR products were analyzed by conformation sensitive gel electrophoresis (CSGE).25 PCR products of abnormal size were submitted to bi-directional sequencing using the ABI Big Dye Terminator Sequencing Kit and detection on an ABI3100 automated capillary sequencer (Applied Biosystems, Foster city, CA). DNA samples with one AG or two AG dinucleotide deletions were analyzed as positive controls for detection of abnormal size of exon 4. A cohort of 340 control European females, 27 Pakistani males and 29 Pakistani females, 11 Turkish males and six Turkish females of normal intelligence were tested for the presence of size variations of PQBP1 exon 4, following the same procedures. DHPLC analyzes of exon 4 PCR fragments from males and females carrying the 21 bp deletion were performed on a WAVE Nucleic Acid Fragment Analysis System (Transgenomic, Inc), at the optimal melting temperatures (Tm) of 63 and 64°C determined using the WaveMaker Software 3.3.3 (Trangenomics, Inc.). X-chromosome inactivation pattern was determined in leucocytes using the human androgen-receptor locus (AR)26 and the FRAXA locus27 as previously reported.
Results
Mentally retarded males were analyzed for mutations in exon 4 of the PQBP1 gene by either fluorescent PCR that can detect insertion or deletion of one base pair or more or by non-fluorescent PCR followed by conformation sensitive gel electrophoresis (CSGE). We found novel deletions in five unrelated patients (Figure 1, Table 1). One patient carried a 23 bp deletion leading to a frameshift (Figure 2). Four unrelated patients carried two different 21 bp in-frame deletions affecting a region coding for five imperfect seven amino-acid tandem repeats in the PRD domain (Figure 3). Three of these deletions are identical and remove the second amino-acid repeat while the third deletion removes the last amino-acid repeat. However, given the redundancy of this region, the proximal deletion can be interpreted in different ways (Figure 3).
In-frame deletions of 21 bp in a region of five imperfect seven amino-acid tandem repeats in the PRD domain (a) Nucleotidic and amino-acid sequences of the 21 bp deletions. The five imperfect seven-amino-acid tandem repeats are underlined. The c.334–354 del(21 bp) (RGHDKSD) and the c.393–413 del(21 bp) (DRGHDKS) are represented in bold. Given the high similarity of the first and second repeat the proximal deletion can also be interpreted in a deletion from c.319–339 del(21 bp). (b) Amino-acid sequence alignment of the human, mouse and rat repeats. Identities are represented by a gray background, conservative changes by an asterisk.
The 23 bp deletion in exon 4 of the PQBP1 gene (c.547–569 del(23 bp)) was found in a patient (patient 1, family 1) of French origin initially referred for systematic screening of fragile X syndrome. This deletion causes a frameshift leading to eight abnormal residues and a premature stop codon E183fsX191. Clinical description was sent to us after mutation identification. The patient walked at the age of 15 months. He stopped normal school education at the age of 7 years. At the age of 9.5 years, neuropsychological evaluation showed hyperactivity, attention deficiency, autistic features and global psychomotor delay (level estimated to be below the age of 4 years). At the age of 10 years when he was seen for the last time (in 2003, prior to mutation identification) his clinical examination showed a global developmental delay associated with hyperactivity, microcephaly (−4SD), short stature (−2SD), clinodactyly, and X-ray showed a delayed bone age and an underdeveloped coccyx bone. Brain imaging (MRI) did not show any associated parenchymal abnormality. Clinical history revealed that his 16 year-old maternal half-brother was mildly mentally retarded, with normal speech but difficulties in reading and writing. He presented with microcephaly (−4SD), short stature (−2SD), facial dysmorphism with prognathism, and prominent ears, nasal speech, bilateral choanal atresia and anal imperforation. Further molecular analysis showed that this half-brother carries the 23 bp deletion and his mother is a heterozygous carrier. The mother is not mentally impaired and presents with partially skewed X-inactivation ratio of 79:21 that is however within the normal range (<80:20). She had four spontaneous miscarriages. Family history did not reveal any other mentally retarded relative. As the mother was pregnant, prenatal diagnosis could be performed to screen for the 23 bp deletion in chorionic villus sample. The male fetus inherited the normal PQBP1 allele (Figure 2).
A 21 bp in-frame deletion (c.334–354 del(21 bp), RGHDKSD motif) was found in exon 4 in three patients: two of the 772 patients of the clinically unselected cohort (patients 2 and 3) and in one of the 17 patients selected on the basis of familial history compatible with XLMR transmission (patient 4).
Patient 2 was first seen at the age of 8 years for borderline MR with attention deficit hyperactivity disorder (ADHD). Parents were consanguineous of Turkish origin, family history was negative (Figure 1, family 2). He did not present with any speech delay. Hyperactivity and attention deficit were first noted at the age of 2.5 years. Neuropsychological evaluation (WISC-III) at 8 years indicated a verbal IQ of 72, a performance IQ of 78 and a global IQ of 72. He presented with behavior anomalies, possibly of sociocultural origin. EEG showed some bilateral occipital paroxystic abnormalities. At the age of 9 years, he was 1-year behind at school, but speech, reading and writing were correct. His height and weight (149 cm, 40 kg), and head circumference (53.5 cm) were normal. He had no dysmorphic feature, but presented with cryptorchidia. Brain MRI showed some bilateral parieto-occipital myelinization delay. His mother, who did not present MR, is a heterozygous carrier for the 21 bp deletion. X-inactivation analysis showed a partial skewing, but the ratio 77:23 was within the normal range.
The same 21 bp in-frame deletion was found in patient 3, born to non-consanguineous parents of Indian origin (Figure 1, family 3). Patient 3 presented with defective speech, behavior anomalies and a typical Fragile X-like phenotype including large and prominent ears, macro-orchidism, hyperactivity and short attention span. At the age of 11 years, a neuropsychological test revealed reduced motor development (corresponding to 7 years), numerical reasoning skills (corresponding to 9 years), and social intelligence (corresponding to 6 years). At age 14 years his height and weight were in the normal lower range (156 cm, 48 kg, both 25 percentile) and head circumference was normal (56 cm 75th centile). Further investigations revealed that his mother and also his grandmother are heterozygous carriers of the same deletion. His maternal uncle inherited the non-deleted gene and is normal.
Patient 4 was 19 years old when last examined (before the mutation identification), born to consanguineous parents (first degree relatives) of Pakistani origin (Figure 1, family 4). He presented with a severe global developmental delay. He walked at the age of 2 years, and he did not learn to read, write or count. He also presented with stereotyped behavior. Neurological examination did not show any neurological abnormality. He presented with borderline short stature (163 cm, −1.5SD) and weighed 49 kg (−2SD). He had a normal head circumference (55 cm) and no dysmorphic features but a rather long face. He has a sister and two brothers, one of whom is 26 years old and is reported to be mentally retarded, but could not be examined and tested for the mutation. His mother was also reported to be mildly retarded.
Another 21 bp deletion affecting a similar amino-acid repeat (c.393–413del(21 bp)), DRGHDKS motif) was found in patient 5, one of the 40 patients selected for the presence of clinical phenotype. This French patient was born to healthy non-consanguineous parents (Figure 1, family 5), after an uneventful pregnancy. At birth, he presented with respiratory distress, requiring cardiac assistance but no assisted ventilation. He showed an early motor delay with walking at 19 months and mild lower limb spasticity. A global psychomotor delay was further observed particularly on speech learning. He was initially evaluated at the age of 2 years and 9 months and showed a global developmental delay without speech, in the absence of autistic features. Dysmorphic features were noted, associating microcephaly (head circumference of 46 cm, −2SD), long ears, small and disjoined teeth, permanently opened mouth with lingual protrusion, clinodactyly of the 5th digit and metatarsus varus of the right foot. The patient had also cryptorchidia. No family history of MR was reported. Brain MRI showed periventricular leucomalacia compatible with his perinatal hypoxia history. Neuropsychological evaluation (Mac-Carthy scale) at the age of 4 years 2 months showed a performance level lower than 2.5 years of age. We analyzed the segregation of the 21 bp deletion. The mother is a heterozygous carrier. She has random X-inactivation in leucocytes (ratio of 26:74). We found the 21 bp deletion in the brother, initially reported as not having developmental delay. Further neuropsychological evaluation (WISC III) at the age of 4.9 years showed intellectual performances in the low normal range, with performance IQ of 88, verbal IQ of 79 and total IQ of 81. Clinical examination did not reveal any malformations or dysmorphic features.
Analysis of 680 control X chromosomes from European females did not reveal any abnormality in PQBP1 the size of exon 4. As no deletion had been detected in a previous dHPLC analysis of exon 4 in 500 European control Xs,5 this indicates that such deletions are very rare in this population (0 in 1180 Xs). As the 334–354del(21 bp) deletion was observed in three non-European patients, we then analyzed control individuals from Pakistani (85 X chromosomes), Turkish (23 Xs) and Indian origin (477 Xs) and found two normal Indian males who carry this c.334–354 del(21 bp) deletion. One male is 44 years old and the other is 22 years old. They both were of normal intelligence without any behavioral abnormalities.
Discussion
In our series of 829 males with MR, we did not find the AG dinucleotide insertion or deletion described previously in five out of 11 families with mutations in PQBP1. 5 We have however found larger deletions, one of 23 bp leading to a frameshift and four of 21 bp that result in loss of one of five copies of an imperfect seven-amino-acid tandem repeat in the PRD domain.
The 23 bp deletion in exon 4 is located after the DR/ER repeat and leads to a truncated PQPB1 protein lacking the NLS signal and the C2 domain. The clinical characteristics of the affected patient and of his half-brother carrying the same deletion are similar to the previously described clinical signs associated with PQBP1 mutations, such as microcephaly, short stature and anal atresia.5, 7, 9
The finding of less disruptive in-frame 21 bp deletions raised the problem of their pathogenic significance. Of the three patients with the (334–354 del(21 bp)) deletion, none presented with microcephaly and one had borderline stature (patient 4). Patient 4 presented a global and severe mental impairment but the two others (patients 2 and 3) had borderline MR and mostly behavioral anomalies. Interfamilial variability has been previously described for some PQBP1 mutations, such as the initially reported AGAG deletion in exon 4, which leads to mild MR and almost no associated features in the MRX55 family, whereas in another family (N45), patients presented with severe MR, microcephaly, short stature and anal atresia.5 The fact that two families were consanguineous would be more suggestive of an autosomal recessive mode of inheritance, but does not necessarily exclude an X-linked trait. In one of them, a brother was reported to have mental impairment; searching for the 21 bp deletion in this case, as for the unaffected male relatives, would be of interest to test association of the 21 bp deletion with MR, but these individuals were not available for clinical and molecular analysis.
The patient with the c.393–413del(21 bp) (patient 5) presented with specific signs such as microcephaly and lower limbs spasticity associated with global and severe mental impairment. Subsequent familial analysis found the 21 bp deletion in a brother initially reported to have normal intelligence, but neuropsychological re-evaluation showed borderline intellectual performances, suggesting variable penetrance or expressivity of the 21 bp deletion. Intrafamilial variability has been described in other families with PQBP1 mutations, with affected males showing profound MR to borderline intelligence.8, 9
The functional relevance of such in-frame deletions can be evaluated from consideration of the position in the domain structure of the protein and conservation in evolution of the target amino-acid sequence. The 21 bp deletions affect a region of five imperfect amino-acid tandem repeats in the PRD domain, present in the predominant isoform of the protein. These deletions would thus affect the predominant isoform but not some of the rare ones. Considering amino-acid conservation during evolution, there are also five repeats of seven aminoacids in mouse and rat, but not in more distant non-mammalian species. Between man and mouse, conservation of these repeats is lower (22/35 identical, 29/35 homologous) than that of the entire proteins (232/265 identical, 251/265 homologous) (Figure 3b). These observations suggest that deletion of one seven amino-acid repeat would not likely have a drastic functional effect. On the other hand, the predicted effects of these changes at the protein level do not exclude an effect on alternative splicing, even though this is not predicted by splice sites scoring programs (http://l25.itba.mi.cnr.it/~webgene/www.spliceview.html), nor by predictive programs of potential exonic splicing enhancers (http://genes.mit.edu/burgelab/rescue-ese).
The 21 bp deletions found in probands with MR (4/829) were initially not observed in a control European population (0/1180 X chromosomes). As we found the c.334–354del(21 bp) deletion in patients from Turkish, Pakistani and Indian origins, we included 23, 85 and 477 control X chromosomes of respectively matched populations. We found the same 21 bp deletion in two Indian controls. This illustrates the importance to analyze large samples of ethnically matched controls when testing the pathogenic nature of putative mutations. Therefore, the c.334–354del(21 bp) deletion appears to be a rare, probably non-pathogenic variant, that may be more frequent among Indian, Pakistani and Turkish individuals than in Europeans. The frequency appears identical in Indian probands 1/254 and Indian controls 2/477. However, the fact that the c.334–354del21(bp) PQBP1 deletion was observed twice in the very small number of non-European, non-Indian patients analyzed leaves open the possibility that this deletion is more frequent in patients than controls. Similarly, a 21 bp deletion was identified in the MEF2A gene from patients affected by an autosomal dominant form of coronary artery disease (CAD) and was proposed to be pathogenic;28 further studies revealed that this deletion was also present in elderly control subjects and did not cosegregate in families with CAD.29
Taking into account all of these data, the c.393–413del(21 bp) deletion might be pathogenic or could at least, as the c.334–354del(21 bp) deletion, act as a low penetrance factor predisposing to mental impairment, by affecting cognitive performances in a subtle way. The recent observation that a recurrent mutation in the ARX gene (24 bp duplication) associated with both syndromic and non-syndromic XLMR is much rarer in sporadic MR male patients than expected from its rather high frequency in families with established XLMR suggested that monogenic XLMR is unlikely to account for the 20–25% of males with MR that would explain the male excess of MR.4 Two additional hypotheses were proposed to account for part of the MR male excess: gender differences in fetal brain development with higher susceptibility of male brain to early brain damage,30 and/or gene polymorphisms on the X chromosome that would subtly affect cognitive abilities and could result in MR when associated with other genetic traits or environmental conditions.4 Further studies in larger cohorts of males with various degrees of cognitive performances would be necessary to establish whether the 21 bp deletions affect MR risk in males.
Our study suggests that truncating mutations in PQBP1 exon 4 are very rare in patients with MR lacking additional evocative clinical signs (see however family MRX555, 13). This may justify clinical selection of patients with family history compatible with XLMR or microcephaly and/or short stature, at least to search for truncating mutations. These signs are quite frequently found in patients with MR, as shown in a South Carolina study of 4008 males with MR. Of these, 12% had microcephaly, 9% had short stature, and 3% had both (cited in6). However, this does not exclude that mutations of milder effect, such as missense mutations, could lead to nonspecific and milder MR. This study also illustrates a common dilemma in XLMR, that is, when missense or in frame rearrangements are found, how to distinguish between mutations and variants that may be non-pathogenic or represent risk factors for cognitive impairment.
References
Stevenson RE, Schroer RJ : X-linked MR. Oxford: Oxford University Press, 2000.
Frints SG, Froyen G, Marynen P, Fryns JP : X-linked MR: vanishing boundaries between non-specific (MRX) and syndromic (MRXS) forms. Clin Genet 2002; 62: 423–432.
Chelly J, Mandel JL : Monogenic causes of X-linked MR. Nat Rev Genet 2001; 2: 669–680.
Mandel JL, Chelly J : Monogenic X-linked MR: is it as frequent as currently estimated? The paradox of the ARX (Aristaless X) mutations. Eur J Hum Genet 2004; 12: 689–693.
Kalscheuer VM, Freude K, Musante L et al: Mutations in the polyglutamine binding protein 1 gene cause X-linked MR. Nat Genet 2003; 35: 313–315.
Lenski C, Abidi F, Meindl A et al: Novel truncating mutations in the polyglutamine tract binding protein 1 gene (PQBP1) cause Renpenning syndrome and X-linked MR in another family with microcephaly. Am J Hum Genet 2004; 74: 777–780.
Stevenson RE, Bennet CW, Abidi F et al: Renpenning syndrome comes into focus. Am J Med Genet 2005; 134A: 415–421.
Fichera M, Falco M, Lo Giudice M et al: Skewed X-inactivation in a family with MR and PQBP1 gene mutation. Clin Genet 2005; 67: 446–447.
Kleefstra T, Franken CE, Arens YH et al: Genotype–phenotype studies in three families with mutations in the polyglutamine-binding protein 1 gene (PQBP1). Clin Genet 2004; 66: 318–326.
Renpenning H, Gerrard JW, Zaleski WA, Tabata T : Familial sex-linked MR. Can Med Assoc J 1962; 87: 954–957.
Sutherland GR, Gedeon AK, Haan EA, Woodroffe P, Mulley JC : Linkage studies with the gene for an X-linked syndrome of MR, microcephaly and spastic diplegia (MRX2). Am J Med Genet 1988; 30: 493–508.
Hamel BC, Mariman EC, van Beersum SE, Schoonbrood-Lenssen AM, Ropers HH : MR, congenital heart defect, cleft palate, short stature, and facial anomalies: a new X-linked multiple congenital anomalies/MR syndrome, clinical description and molecular studies. Am J Med Genet 1994; 51: 591–597.
Deqaqi SC, N'Guessan M, Forner J et al: A gene for non-specific X-linked MR (MRX55) is located in Xp11. Ann Genet 1998; 41: 11–16.
Iwamoto K, Huang Y, Ueda S : Genomic organization and alternative transcripts of the human PQBP-1 gene. Gene 2000; 259: 69–73.
Imafuku I, Waragai M, Takeuchi S et al: Polar amino acid-rich sequences bind to polyglutamine tracts. Biochem Biophys Res Commun 1998; 25: 16–20.
Waragai M, Lammers CH, Takeuchi S et al: PQBP-1, a novel polyglutamine tract-binding protein, inhibits transcription activation by Brn-2 and affects cell survival. Hum Mol Genet 1999; 8: 977–987.
Okazawa H, Rich T, Chang A et al: Interaction between mutant ataxin-1 and PQBP-1 affects transcription and cell death. Neuron 2002; 34: 701–713.
Komuro A, Saeki M, Kato S : Association of two nuclear proteins, Npw38 and NpwBP, via the interaction between the WW domain and a novel proline-rich motif containing glycine and arginine. J Biol Chem 1999; 274: 36513–36519.
Okazawa H, Sudol M, Rich T : PQBP-1 (Np/PQ): a polyglutamine tract-binding and nuclear inclusion-forming protein. Brain Res Bull 2001; 56: 273–280.
Komuro A, Saeki M, Kato S : Npw38, a novel nuclear protein possessing a WW domain capable of activating basal transcription. Nucleic Acids Res 1999; 2: 1957–1965.
Craggs G, Finan PM, Lawson D et al: A nuclear SH3 domain-binding protein that colocalizes with mRNA splicing factors and intermediate filament-containing perinuclear networks. J Biol Chem 2001; 276: 30552–30560.
Llorian M, Beullens M, Andres I et al: SIPP1, a novel pre-mRNA splicing factor and interactor of protein phosphatase-1. Biochem J 2004; 378: 229–238.
Waragai M, Junn E, Kajikawa M et al: PQBP-1/Npw38, a nuclear protein binding to the polyglutamine tract, interacts with U5–15kD/dim1p via the carboxyl-terminal domain. Biochem Biophys Res Commun 2000; 273: 592–595.
Reuter K, Nottrott S, Fabrizio P, Lührmann R, Ficner R : Identification, characterization and crystal structure analysis of the human spliceosomal U5 snRNP-specific 15 kD protein. J Mol Biol 1999; 294: 515–525.
Ganguly A, Rock MJ, Prockop DJ : Conformation sensitive gel electrophoresis for rapid detection of single base differences in double-stranded PCR products and DNA fragments: evidence for solvent induced bends in DNA heteroduplexes. Proc Natl Acad Sci USA 1993; 90: 10325–10329.
Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW : Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 1992; 5: 1229–1239.
Carrel L, Willard HF : An assay for X inactivation based on differential methylation at the fragile X locus, FMR1. Am J Med Genet 1996; 64: 27–30.
Wang L, Fan C, Topol SE, Topol EJ, Wang Q : Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science 2003; 302: 1578–1581.
Weng L, Kavaslar N, Ustaszewska A et al: Lack of MEF2A mutations in coronary artery disease. J Clin Invest 2005; 115: 1016–1020.
de Courten-Myers GM : The human cerebral cortex: gender differences in structure and function. J Neuropathol Exp Neurol 1999; 58: 217–226.
Acknowledgements
We wish to acknowledge the financial support of the Ministère de la santé, de la famille et des personnes handicapées; DST-DAAD, Deutsche Forschungsgemeinschaft; and DBT grant #BT/PR4959/Med/14/375/2004. We thank Mrs Kindelberger for contribution to neuropsychological evaluation and Dr Mitesh Shetty of Manipal Hospital, Bangalore for valuable clinical inputs.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cossée, M., Demeer, B., Blanchet, P. et al. Exonic microdeletions in the X-linked PQBP1 gene in mentally retarded patients: a pathogenic mutation and in-frame deletions of uncertain effect. Eur J Hum Genet 14, 418–425 (2006). https://doi.org/10.1038/sj.ejhg.5201593
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201593
Keywords
This article is cited by
-
Changes in the folding landscape of the WW domain provide a molecular mechanism for an inherited genetic syndrome
Scientific Reports (2016)
-
Mutations in the PQBP1 gene prevent its interaction with the spliceosomal protein U5–15kD
Nature Communications (2014)
-
Comparative Genetics of the Poly-Q Tract of Ataxin-1 and Its Binding Protein PQBP-1
Biochemical Genetics (2012)
-
A two base pair deletion in the PQBP1 gene is associated with microphthalmia, microcephaly, and mental retardation
European Journal of Human Genetics (2007)