Abstract
Hereditary congenital facial palsy (HCFP) is an autosomal–dominant disorder consisting of paresis or paralysis of the VIIth (facial) cranial nerve. Genetic heterogeneity for this disorder has been suggested based on linkage analysis in two large Dutch families. Two loci have been identified, one on chromosome 3q21.2–q22.1 (HCFP1) and another on chromosome 10q21.3–q22.1 (HCFP2). Here, we report linkage analysis in a large Pakistani family with dominant congenital facial palsy. A region cosegregating with the disorder was identified on the long arm of chromosome 3, which overlaps with the previously identified HCFP1 locus on chromosome 3q21–q22, thus confirming the involvement of this locus in HCFP. The critical region could be reduced from 5.7 to 3.0 cM between the markers D3S3607 and GDB ID:11524500. In addition, mutation analysis on seven candidate genes: KLF15, FLJ40083, PODXL2, TMCC1, PLEXIN-A1, PLEXIN-D1, and GATA-2, was performed. All genes are located within the critical interval of the Dutch HCFP1 family. The genes PODXL2, PLEXIN-D1, GATA-2, and TMCC1 are also located within the smaller critical interval of the Pakistani HCFP family. Based on the results obtained, all seven genes could be excluded as causative genes in HCFP.
Similar content being viewed by others
Introduction
Congenital facial palsy belongs to the group of congenital diseases characterized by abnormal eye, eyelid, and/or facial movement, referred to as congenital cranial dysinnervation disorders or in short CCDDs.1 Disorders belonging to the CCDDs include Duane syndrome, Möbius syndrome, horizontal gaze palsy, congenital ptosis, and congenital facial palsy. To date, genetic defects underlying several of these syndromes have been identified, including KLF21A and PHOX2A involved in congenital fibrosis of the extraocular muscles (CFEOM), CPAH and SALL4 involved in Duane's retraction syndrome, and ROBO3 involved in horizontal gaze palsy and progressive scoliosis.2, 3, 4, 5, 6, 7, 8, 9, 10
For Möbius syndrome (MIM157900) and hereditary congenital facial nerve palsy (HCFP; MIM601471, MIM604185), loci have been identified,11, 12, 13, 14, 15, 16 but the genetic defects underlying these disorders are still unknown. Although both disorders belong to the CCDDs and share some of the same clinical features, they are considered different entities based on recent clinical and neuropathological findings.17 Möbius syndrome is considered a developmental disorder of the entire lower brainstem and minimal diagnostic criteria are congenital facial weakness with impairment of minimally the facial (nVII) and the abducens nerve (nVI) and possible involvement of other cranial nerves, craniofacial dysmorphisms, limb malformations, and long tract involvement.18 Based on chromosomal aberrations, chromosome 13q12.2–q13 was identified as a locus involved in Möbius syndrome and a second possible locus has been identified on chromosome 1p22.11, 13, 14, 16 A third possible locus has been postulated based on chromosome 22q11 deletions in three unrelated patients with among other clinical features also congenital unilateral facial palsy.19
Congenital facial weakness is proposed to result from maldevelopment of the facial nucleus and/or cranial nerve and has been defined as complete or partial absence of the facial nerve (nVII), either uni- or bilateral.17 Several small and two large HCFP families (HCFP1 and HCFP2) have been described. The phenotype described for the HCFP1 (MIM601471, formerly known as MBS2) family is an asymmetric, mostly bilateral, weakness of the facial muscles and an unequal involvement of the three branches of the facial nerve. Inheritance is autosomal dominant with a penetrance of 95%. Linkage analysis identified a locus on chromosome 3q21–q22 cosegregating with the disorder.12, 20 Several candidate genes with a possible function in hindbrain development or with an expression pattern suggesting a role during hindbrain patterning, such as PLEXIN-D1, PLEXIN-A1, and GATA-2, were screened in order to identify the genetic defect underlying HCFP1, but no causative mutations have been identified.20, 21, 22 The phenotype of affected members of the HCFP2 (MIM604185, formerly known as MBS3) family is slightly different from the HCFP1 phenotype and is characterized as an often asymmetrical, uni- or bilateral, facial weakness, and an unequal involvement of the three branches of the facial nerve. Besides facial weakness, hearing loss and congenital deafness were reported. Inheritance is autosomal dominant with a penetrance of 60%. Linkage analysis identified a locus on chromosome 10q21.3–q22.1 cosegregating with the disorder.15 Mutation analysis on the candidate genes EGR2, CTNNA3, and LRRTM3 located within or nearby the critical region failed to identify the underlying genetic defect for HCFP2 (B van der Zwaag, unpublished data).20 For the smaller HCFP families, linkage analysis was not reported. Identification of multiple loci for congenital facial palsy suggests genetic heterogeneity for this disorder.
In this paper, we describe a large Pakistani family with autosomal–dominant congenital facial palsy that, like the Dutch HCFP1 family, maps to the long arm of chromosome 3. The critical region could be reduced to 3.0 cM (∼3.0 Mb). Furthermore, mutation analysis was performed on candidate genes located within the redefined critical region for HCFP1.
Materials and methods
Patients
In total, 29 blood samples were collected from members of the Pakistani family, a part of which lives in the UK and the others in Pakistan. Neurological assessment of the proband (IV.13; Figure 1) led to the diagnosis of unilateral lower motor neuron palsy of the VIIth cranial nerve. Further neurological examination, CT and MRI scan, appeared to be normal and did not show any facial nucleus problems or any other focal lesions. Genetic investigations excluded chromosomal rearrangements and no evidence was obtained for chromosome 22q11 microdeletion. DNA studies for myotonic dystrophy and facioscapulohumeral dystrophy were normal.
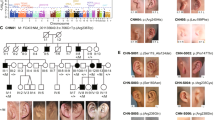
Pedigree of the family and haplotypes of individuals available for this study. The ancestral mutation-bearing chromosome is boxed. The line besides the haplotypes marks the part of the ancestral chromosome present in the patients. Deduced haplotypes are given in brackets. The borders of the critical linkage interval were proximally defined by individual IV.12 and IV.13 and distally defined by individual V.4. In addition, a null allele was observed for marker GDB:11524499 in the individuals IV.3, V.2, and V.3. The markers D3S1551, D3S1589, D3S1541, ACPP, and D3S1290 were omitted from the figure for clarity.
Genotyping
Genomic DNA used for typing of the DNA polymorphisms was isolated as described by Miller et al.23 Amplification of the polymorphic regions and analysis of the amplified fragments were performed according to Kremer et al.24 Polymorphic markers from the 3q21 region included D3S1551, D3S1589, D3S3607, RHO, D3S1587, D3S3548, D3S3514, D3S1292, D3S1541, ACPP, and D3S1290. The order of the markers is according to the Human Genome Working Draft (May 2004; http://genome.ucsc.edu). Primers for five new CA-repeat markers around the RHO marker were designed using the Human Genome Working Draft (May 2004; simple repeat set at full) followed by in silico PCR analysis (see Table 1 for primers and amplicon length).
Linkage analysis
Two-point lod scores and the maximum lod score were calculated with the EasyLinkagePlus program, subroutine SuperLink V1.4-Two-Point Parametric Linkage Analysis.25, 26 The disease allele frequency was set at 0.0001 and a penetrance of 95% was assumed.
Candidate gene analysis
The candidate gene analysis and the multiplex PCR were performed on genomic DNA derived from individuals IV.6, IV.7, and V.17 from the Pakistani HCFP1 family and from individuals V.3, V.4, and IV.13 from the Dutch HCFP1 family.12 PCR products covering the coding regions, intron–exon boundaries, and 5′ untranslated regions of PLEXIN-A1 (GenBank ID: NM_032242), GATA-2 (GenBank ID: NM_032638), PLEXIN-D1 (GenBank ID: NM_015103), KLF15 (GenBank ID: NM_014079), FLJ40083 (GenBank ID: NM_182628), PODXL2 (GenBank ID: NM_015720), and TMCC1 (GenBank ID: NM_001017395/NM_015008) were generated. Primer sequences and conditions for amplification are available upon request. The PCR products were purified with QIAquick Gel Extraction Kit (Qiagen). Direct sequencing was performed using the ABI PRISM BigDye Terminator Cycle Sequencing V2.0 Ready Reaction Kit and analyzed with the ABI PRISM 3730 DNA Analyzer (Applied Biosystems). Multiplex ligation-dependent probe amplification (MLPA) with minimally two probes per gene was performed as described (www.mrc-holland.com). Probe sequences are available upon request.
Results
Linkage analysis
A Pakistani family was diagnosed with autosomal–dominant HCFP, marked as unilateral facial palsy of the VIIth cranial nerve. The phenotype of the Pakistani family presented is this work resembles the phenotype described for the Dutch HCFP1 (MIM601471; retired MBS2) family, in which linkage to chromosome 3q21–22 was identified.12 Therefore, linkage analysis in the Pakistani family was carried out with 11 polymorphic markers and an additional five CA-repeat markers located within the chromosome 3q21–22 interval. Twenty-nine persons were included in the DNA analysis, 19 of whom were affected.
The order of the markers tested within the Dutch HCFP1 family12, 20 was reordered according to the Human Genome Working Draft (May 2004) and additional markers located within the critical interval of this family were tested (data not shown), leading to a redefinition of the critical region for the HCFP1 locus (previously: D3S1589-ACPP20). According to our current knowledge, the critical linkage interval for the HCFP1 family is defined as the region D3S1589;D3S3514 spanning 5.64 Mb (5.7 cM, according to the deCODE genetic map).27
Linkage analysis in the Pakistani family showed that this family, like the Dutch HCFP1 family, maps to the long arm of chromosome 3. Two-point lod scores are given in Table 2. The highest lod score was 6.90 at θ=0.00 for marker GDB ID:11524498. For calculation of the lod scores family member V.1 was excluded. Although this individual carries the at-risk haplotype, she enters the family via another branch without affected individuals and her connection to the other affected individuals is unclear.
Haplotypes were constructed to define the borders of the cosegregating region. The at-risk haplotype in the 3q21 region is depicted in Figure 1. Based on 16 polymorphic markers tested, the borders of the cosegregating region were defined. The proximal border of the critical region is determined by a recombination, present in individuals IV.12 and IV.13 and their affected offspring, between the markers D3S3607 and GDB ID:11524496 (Figure 1). The distal border of the region is determined by a recombination, present in individual V.4, between the markers GDB ID:11524499 and GDB ID:11524500 (Figure 1). The critical linkage interval within this family was defined as region D3S3607;GDB ID:11524500 spanning 3.0 Mb (3.0 cM, according to the deCODE genetic map).27 In contrast to the other described HCFP families, there are no obligate carriers or unaffected members carrying the at–risk haplotype, leading to a penetrance of 100% in this family.
The at-risk haplotypes from the Dutch and the Pakistani HCFP1 families were clearly different (data not shown), ruling out the remote possibility of a common founder. The at-risk haplotype of the Pakistani family reduced the critical region on chromosome 3q21 known from the Dutch HCFP1 family on which a genetic defect involved in facial palsy is located by 47% (from 5.7 to 3.0 cM).
Candidate gene mutation analysis
A genome database survey of the critical linked interval revealed that the region contains 35 and 54 predicted transcripts for the interval of the Pakistani HCFP1 and the Dutch HCFP1 family, respectively (Human Genome Working Draft, May 2004). Several candidate genes were selected based on their cellular function and/or based on their expression pattern during (hind-) brain development and tested in both families. Four candidate genes located within the critical interval of the Dutch HCFP1 family: KLF15, FLJ40083, PODXL2, and TMCC1 were indicated as putative candidate genes based on their expression pattern observed during hindbrain development, using mouse RNA in situ hybridization.28 Two of these genes, PODXL2 and TMCC1, are also located within the critical interval of the Pakistani family (Figure 2). KLF15, a member of the Krüppel-like protein family of transcriptional regulators, is expressed in the neuroepithelium of the developing mouse brain and is implicated in a large number of processes, including the differentiation of neuronal precursors.29 FLJ40083, a predicted protein with no homology to known proteins, has a large overlapping expression pattern with KLF15 during mouse brain development. PODXL2, coding for an endoglycan protein, a member of the CD34/podocalyxin family of sialomucins, is highly expressed in the central and peripheral nervous system. Highest levels of expression were observed in dorsal cells of the cerebellar primordium, the hindbrain, and anterior part of the medulla oblongata and spinal cord. The last selected candidate gene, TMCC1, has moderate homology to the plectin proteins, which are important for cell stability, and interact with the actin cytoskeleton in cell remodeling.30 TMCC1 expression was observed in the brain, spinal cord, and all ganglia. Furthermore, the candidate genes PLEXIN-A1, GATA-2, and PLEXIN-D1, which were previously tested in the Dutch HCFP1 family,20, 21, 22 were also tested in the Pakistani HCFP1 family. Based on the putative function and expression pattern of these three genes, they remain candidate genes for HCFP1. PLEXIN-A1 and PLEXIN-D1 are both members of the plexin family of transmembrane receptors, which are implicated in axonal guidance and cell–cell contact.31, 32 During mouse embryogenesis, PLEXIN-D1 is expressed in the central and peripheral nervous system, and in the vascular endothelium of developing blood vessels.33 PLEXIN-A1 is expressed in the forebrain, midbrain, hindbrain, spinal cord, and dorsal root ganglia.21 Finally, GATA-2, a member of the GATA-binding protein family of transcription factors, is expressed in the developing hindbrain, with its expression limited to rhombomere 4, an important structure for the development of the facial nerve.34 PCR amplicons covering the entire coding sequence and the intron–exon boundaries of the seven candidate genes were generated. Mutation analysis was carried out by direct sequencing of amplicons derived from individuals IV.6, IV.7, and V.17 for the Pakistani family and individuals V.3, V.4, and IV.13 for the Dutch family.12 Analysis of TMCC1, FLJ40083, PODXL2, GATA-2, PLEXIN-D1, and PLEXIN-A1 revealed three, four, five, five, 23, and 19 nucleotide changes, respectively (see Supplementary data). The observed variations were unlikely to be causative mutations, as they were either already known single–nucleotide polymorphisms or did not cosegregate with the disorder. For KLF15, no nucleotide changes were identified in either family. As the candidate genes PODXL2, GATA-2, PLEXIN-D1, and TMCC1 were the only remaining candidate genes within the critical region identified in the Pakistani HCFP1 family, we also analyzed the first 1000 bp upstream of the start codon for PODXL2, GATA-2, and PLEXIN-D1. TMCC1 was excluded from this analysis, as it was difficult to define regulatory sequences for this gene. In the case of PODXL2 and PLEXIN-D1, additional polymorphisms were identified (see Supplementary data). To exclude gene deletion (in case of KLF15) or duplication, MLPA on all seven candidate genes was performed. No aberrant copy number of the candidate genes was identified (data not shown). Although known and unknown polymorphisms were identified, neither a causative mutation cosegregating with the disorder nor a gene duplication or deletion was identified in either family, making it unlikely that one of the seven genes screened is involved in HCFP.
Discussion
HCFP, a dominant inherited autosomal disorder, has been described for two Dutch families HCFP1 (MIM601471) and HCFP2 (MIM604185). Linkage analysis in these two families identified two different loci cosegregating with the disorder, chromosome 3q21–22 in HCFP1 and chromosome 10q21–22 in HCFP2,12, 15 indicating genetic heterogeneity for this disorder.
In the present study, we describe a third large HCFP family originating from Pakistan. Linkage analysis identified a region cosegregating with the disorder on the long arm of chromosome 3, colocalizing to the identified HCFP1 3q21–3q22 interval, and confirmed the involvement of this locus in congenital facial palsy. The linkage intervals of the Dutch HCFP1 and the Pakistani HCFP1 families were compared to each other. First of all, using the current physical map of chromosome 3 and testing additional polymorphic markers in the Dutch HCFP1 family, the critical region in this family was redefined and is delimited by the markers D3S1589 and D3S3514 and spans about 5.7 cM at chromosome 3q21.2–q22.1. The critical region in the Pakistani HCFP1 family is entirely located within the critical region of the Dutch HCFP1 family and is delimited by the markers D3S3607 and GDB ID:11524500 and spans about 3.0 cM at chromosome 3q21.3. These new findings reduced the interval in which the genetic defect underlying congenital facial palsy is located by 47%. The at–risk haplotype in both families is different, ruling out a common founder. The HCFP1 locus contains 54 and 35 predicted transcripts for the Dutch and the Pakistani family, respectively. Seven candidate genes, KLF15, PODXL2, FLJ40083, TMCC1, PLEXIN-A1, PLEXIN-D1, and GATA-2, of which the latter three were screened before in the HCFP1 family,20, 21, 22 were selected based on their expression profile and (predicted) function and screened by mutation analysis in both families. Although numerous DNA variations were identified, none of these were likely to be causative for the HCFP phenotype. At present, we cannot exclude whether expression of these candidate genes is altered, owing to mutations in regulatory sequences, leading to an aberrant expression pattern during hindbrain developmental stages. The temporal and spatial transcriptional control in the development of these genes might be altered; unfortunately, this cannot be assessed in the available patient material.
At the moment, there are no other obvious candidate genes located within the linked interval. Of the 35 putative transcripts located within the D3S3607;GDB ID:11524500, six were ruled out by direct sequencing: PODXL2, GATA-2, TMCC1, PLEXIN-D1, RAB7, and EEFSEC,20, 22, 28 interval, and another four (MBD4, KIAA1257, MCM2, and ABTB1) based on their spatio/temporal expression during development.28 Of the remaining 25 transcripts, five are potentially expressed in the brain (TRH, ACAD9, DNAJB8, GPR175, and KLHDC6); although these genes were tested using RNA in situ hybridization, they were discarded as putative candidate genes owing to lack of a signal during mouse embryogenesis.28 However, the expression levels could be below detection level and therefore it might be worthwhile to screen these genes by mutation analysis. In the course of this study, materials from three smaller HCFP families originating from England, Spain, and The Netherlands were obtained. Polymorphic markers located within the known HCFP1 and HCFP2 loci were tested by linkage analysis in these three small families. In two families, chromosome 10 could be excluded to be involved in the HCFP phenotype; however, a possible linkage to chromosome 3 was observed (data not shown). This suggests that the chromosome 3q21 locus may be an important locus for the HCFP1 disorder. These two smaller families will be included in future candidate gene analysis.
In conclusion, the involvement of chromosome 3q21–q22 in HCFP could be confirmed by linkage analysis in a third large HCFP family and the critical interval cosegregating with the disorder could be reduced by 47% to 3.0 cM on chromosome 3q21.3. Furthermore, the candidate genes KLF15, PODXL2, FLJ40083, TMCC1, PLEXIN-A1, PLEXIN-D1, and GATA-2 can be excluded to be involved in the HCFP1 phenotype.
References
Gutowski NJ, Bosley TM, Engle EC : 110th ENMC International Workshop: the congenital cranial dysinnervation disorders (CCDDs). Neuromuscul Disord 2003; 13: 573–578.
Al Baradie R, Yamada K, St Hilaire C et al: Duane radial ray syndrome (Okihiro syndrome) maps to 20q13 and results from mutations in SALL4, a new member of the SAL family. Am J Hum Genet 2002; 71: 1195–1199.
Butcher J : Mutations in ROBO3 cause HGPPS. Lancet Neurol 2004; 3: 328.
Jen JC, Chan WM, Bosley TM et al: Mutations in a human ROBO gene disrupt hindbrain axon pathway crossing and morphogenesis. Science 2004; 304: 1509–1513.
Kohlhase J, Heinrich M, Schubert L et al: Okihiro syndrome is caused by SALL4 mutations. Hum Mol Genet 2002; 11: 2979–2987.
Nakano M, Yamada K, Fain J et al: Homozygous mutations in ARIX (PHOX2A) result in congenital fibrosis of the extraocular muscles type 2. Nat Genet 2001; 29: 315–320.
Pizzuti A, Calabrese G, Bozzali M et al: A peptidase gene in chromosome 8q is disrupted by a balanced translocation in a Duane syndrome patient. Invest Ophthalmol Vis Sci 2002; 43: 3609–3612.
Yamada K, Andrews C, Chan WM et al: Heterozygous mutations of the kinesin KIF21A in congenital fibrosis of the extraocular muscles type 1 (CFEOM1). Nat Genet 2003; 35: 318–321.
Yamada K, Chan WM, Andrews C et al: Identification of KIF21A mutations as a rare cause of congenital fibrosis of the extraocular muscles type 3 (CFEOM3). Invest Ophthalmol Vis Sci 2004; 45: 2218–2223.
Yamada K, Hunter DG, Andrews C, Engle EC : A novel KIF21A mutation in a patient with congenital fibrosis of the extraocular muscles and Marcus Gunn jaw-winking phenomenon. Arch Ophthalmol 2005; 123: 1254–1259.
Donahue SP, Wenger SL, Steele MW, Gorin MB : Broad-spectrum Möbius syndrome associated with a 1;11 chromosome translocation. Ophthalmic Paediatr Genet 1993; 14: 17–21.
Kremer H, Kuyt LP, van den Helm HB et al: Localization of a gene for Möbius syndrome to chromosome 3q by linkage analysis in a Dutch family. Hum Mol Genet 1996; 5: 1367–1371.
Nishikawa M, Ichiyama T, Hayashi T, Furukawa S : Möbius-like syndrome associated with a 1;2 chromosome translocation. Clin Genet 1997; 51: 122–123.
Slee JJ, Smart RD, Viljoen DL : Deletion of chromosome 13 in Möbius syndrome. J Med Genet 1991; 28: 413–414.
Verzijl HT, van den Helm HB, Veldman B et al: A second gene for autosomal dominant Möbius syndrome is localized to chromosome 10q, in a Dutch family. Am J Hum Genet 1999; 65: 752–756.
Ziter FA, Wiser WC, Robinson A : Three-generation pedigree of a Möbius syndrome variant with chromosome translocation. Arch Neurol 1977; 34: 437–442.
Verzijl HT, van der Zwaag B, Lammens M, ten Donkelaar HJ, Padberg GW : The neuropathology of hereditary congenital facial palsy vs Möbius syndrome. Neurology 2005; 64: 649–653.
Verzijl HT, van der Zwaag B, Cruysberg JR, Padberg GW : Möbius syndrome redefined: a syndrome of rhombencephalic maldevelopment. Neurology 2003; 61: 327–333.
Puñal JE, Siebert MF, Angueira FB, Lorenzo AV, Castro-Gago M : Three new patients with congenital unilateral facial nerve palsy due to chromosome 22q11 deletion. J Child Neurol 2001; 16: 450–452.
van der Zwaag B, Verzijl HT, Beltran-Valero dB et al: Mutation analysis in the candidate Möbius syndrome genes PGT and GATA2 on chromosome 3 and EGR2 on chromosome 10. J Med Genet 2002; 39: E30.
van der Zwaag B, Burbach JP, Brunner HG, van Bokhoven H, Padberg GW : Nucleotide variation analysis does not support a causal role for plexin-A1 in hereditary congenital facial paresis. Brain Res Dev Brain Res 2005; 158: 66–71.
van der Zwaag B, Verzijl HT, Wichers KH et al: Sequence analysis of the PLEXIN-D1 gene in Möbius syndrome patients. Pediatr Neurol 2004; 31: 114–118.
Miller SA, Dykes DD, Polesky HF : A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215.
Kremer H, Pinckers A, van den Helm HB, Deutman AF, Ropers HH, Mariman EC : Localization of the gene for dominant cystoid macular dystrophy on chromosome 7p. Hum Mol Genet 1994; 3: 299–302.
Fishelson M, Geiger D : Exact genetic linkage computations for general pedigrees. Bioinformatics 2002; 18 (Suppl 1): S189–S198.
Lindner TH, Hoffmann K : easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics 2005; 21: 405–407.
Kong A, Gudbjartsson DF, Sainz J et al: A high-resolution recombination map of the human genome. Nat Genet 2002; 31: 241–247.
van der Zwaag B, Burbach JPH, Scharfe C et al: Identifying new candidate genes for hereditary facial paresis on chromosome 3q21–q22 by RNA in situ hybridization in mouse. Genomics 2005; 86: 55–67.
Bieker JJ : Krüppel-like factors: three fingers in many pies. J Biol Chem 2001; 276: 34355–34358.
Allen PG, Shah JV : Brains and brawn: plectin as regulator and reinforcer of the cytoskeleton. Bioessays 1999; 21: 451–454.
Tamagnone L, Artigiani S, Chen H et al: Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell 1999; 99: 71–80.
Tamagnone L, Comoglio PM : Signalling by semaphorin receptors: cell guidance and beyond. Trends Cell Biol 2000; 10: 377–383.
van der Zwaag B, Hellemons AJ, Leenders WP et al: PLEXIN-D1, a novel plexin family member, is expressed in vascular endothelium and the central nervous system during mouse embryogenesis. Dev Dyn 2002; 225: 336–343.
Auclair F, Valdes N, Marchand R : Rhombomere-specific origin of branchial and visceral motoneurons of the facial nerve in the rat embryo. J Comp Neurol 1996; 369: 451–461.
Acknowledgements
We thank the family members for their participation in this study. This work was partially funded by the Prinses Beatrix Fonds project number MAR03-0117 and Stichting Neurologie en Wetenschap.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Michielse, C., Bhat, M., Brady, A. et al. Refinement of the locus for hereditary congenital facial palsy on chromosome 3q21 in two unrelated families and screening of positional candidate genes. Eur J Hum Genet 14, 1306–1312 (2006). https://doi.org/10.1038/sj.ejhg.5201706
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201706
Keywords
This article is cited by
-
A framework for the evaluation of patients with congenital facial weakness
Orphanet Journal of Rare Diseases (2021)