Abstract
A genome-wide linkage analysis of platelet count was carried out in a large Asian Indian kindred. Linkage analysis showed one marker (D3S1309) on chromosome 3q with a lod score of 3.26 and another (D3S1282) approximately 30 cM centromeric, with a lod score of 2.52. Multipoint analysis of chromosome 3q identified two peaks with maximum multipoint lod scores of 3.52 and 4.11 under markers D3S1309 and D3S1282, respectively. Two strong candidate genes for platelet variation were identified in the linked region; thrombopoietin (THPO) and glycoprotein IX (GPIX). Resequencing of four individuals revealed five single-nucleotide polymorphisms (SNPs) in THPO and one mutation in the transmembrane region of GPIX. Analysis of variance showed that the GPIX mutation and one THPO SNP accounted for 6 and 4% of the variation in platelet count, respectively. The THPO SNP lies in the 3′ untranslated region of the gene and has not been previously reported. The G to A transition at nucleotide 653 resulted in an Ala 156 (GCC) to Thr (ACC) replacement in the GPIX protein. The GPIX mutation was recently identified in a Chinese patient with Bernard–Soulier syndrome (BSS), a rare recessive bleeding disorder characterized by thrombocytopenia and giant platelets. One copy of the GPIX mutation was found in 300 European individuals with platelet counts within the normal range. The results suggest that two QTLs on chromosome 3q influence platelet count variation in the Asian Indian kindred, with the GPIX transmembrane mutation and the 3′ UTR SNP in THPO being strong candidates.
Similar content being viewed by others
Introduction
Platelets are small anuclear cells that adhere to damaged blood vessels and effect hemostasis. They contain RNA, a canalicular system, and several different types of granules that play an important role in the formation of blood clots and the inflammatory response. Platelets are produced in the bone marrow from mature megakaryocytes and circulate in blood for 9–10 days before being sequestered in the spleen for elimination. The hemopoietic growth factor, thrombopoietin (TPHO), is the primary regulator of megakaryocyte growth and platelet numbers.1, 2 TPHO is produced primarily in the liver, at a constitutive rate. Plasma TPHO level is regulated in a negative feedback fashion via adsorption and internalization by cells that express the TPHO receptor, Mpl.3, 4, 5, 6, 7, 8, 9 Thus, when platelets and megakaryocytes are decreased, less TPHO is removed from the plasma, the subsequent increase in circulating TPHO stimulates an increase in platelet production in the bone marrow.
Thrombopoiesis, however, is not entirely dependent on TPHO. Other genes that participate in megakaryopoiesis, megakaryocyte differentiation, and the subsequent shedding of platelets, include glycoprotein (GP)11b, GP1bα, GPIX, GPV, platelet factor 4, and the gene for the TPHO receptor, c-Mpl.10 Several clinical disorders of low platelet count (thrombocytopenia) and high platelet count (thrombocytosis) have been documented. While most cases of thrombocytopenia are more likely to be acquired than inherited, several syndromes of congenital thrombocytopenia have been described, including those involving the GP1bα and GPIX genes (Bernard–Soulier syndrome (BSS)).11, 12, 13
Although the genetic determinants of many congenital thrombocytopenias have been identified, very little is known about the genetic determinants of normal platelet count variation. The ‘normal’ range of platelet count in a healthy person is quite broad (150–400 × 109/l), but an individual's platelet count is maintained within a much narrower range; constantly balanced between thrombopoiesis and senescence.14 Platelet count within the normal range has been shown to be largely determined by genetic factors with heritability estimates from twin studies ranging from 0.57 to 0.80.15, 16 The first genome-wide linkage analysis of platelet count was recently reported with 745 Australian twin pairs having platelet counts within the normal range and showed suggestive evidence for a QTL on chromosome 19q (lod score=2.59, P-value=0.0003).17
The results of a genome-wide linkage study of a large Asian Indian kindred with β thalassemia are reported here. Significant linkage is shown at two locations on chromosome 3q with age-, sex-, and β thalassemia-adjusted platelet counts. Analysis of two candidate genes on chromosome 3q is described. A known, rare functional mutation in the platelet glycoprotein IX (GPIX) gene18 was shown to be associated with low platelet counts in the kindred. The mutation creates a hydrophilic amino acid (threonine) from a hydrophobic amino acid (alanine) in the transmembrane domain of the mature protein. A previously unknown single-nucleotide polymorphism (SNP) in the 3′ untranslated region (UTR) of the thrombopoietin gene was shown to be associated with platelet counts in the kindred; although no function could be attributed to the SNP it could potentially influence post-transcriptional control of gene expression.
Materials and methods
Subjects
The kindred of Asian Indian origin used in this study has been previously described in detail.19, 20, 21 β thalassemia and hereditary persistence of fetal hemoglobin (HPFH) segregate within the kindred. Three individuals were homozygous for β thalassemia and they were removed from the analysis. The kindred spans seven generations and was analyzed in six family units in order to break consanguineous relationships and reduce computation time, as described previously.20
The six families include 7, 11, 16, 49, 59, and 75 individuals, with 4, 9, 15, 37, 27, and 62 of the individuals being genotyped, 7, 6, 11, 36, 51, and 62 of the individuals having platelet count data and 4, 6, 11, 31, 25, and 48 individuals having both genotype and platelet count data in each family, respectively.
Hematological studies
Blood samples were collected in EDTA as anticoagulant. Hemoglobin level, red blood cell counts (RBC), white cell counts (WBC), and platelets counts were determined with an automated blood cell analyzer (Bayer H3 RTX, Newbury, UK).
Genotyping
The marker data were the same as used by Craig et al20 in a previous genome screen for genes acting on fetal hemoglobin levels in the kindred; five additional markers were added later under the same protocol. A total of 216 microsatellite markers were genotyped in total on the autosomal chromosomes, and 10 on chromosome X. All identifiable genotyping errors were corrected for the previous analysis of the marker data. The marker allele frequencies were estimated from the data.
Four individuals in the kindred were selected for mutation detection based on their platelet levels; two individuals with platelet levels at least one standard deviation greater than the kindred mean and two with platelet counts at least one standard deviation less than the kindred mean.
DNA analyses
DNA was extracted from peripheral blood leukocytes using standard procedures. Four individuals were selected for sequence analyses of the GPIX and THPO genes. The polymerase chain reaction (PCR) was used to specifically amplify the entire genomic sequence of the GPIX gene (ENSEMBL gene ENSG000001/69704) in five overlapping amplicons, which included three exons and both introns, about 250 bp upstream and 100 bp downstream genomic sequence, respectively. The PCR amplifications were performed in 25 μl reaction volumes containing 50 ng genomic DNA, 0.2 mmol/l of each dNTP, 50 mmol/l KCl, 10 mmol/l Tris-HCl pH 8.3, 1.5 mmol/l MgCl2, 2.0 U Taq Gold polymerase (PE Applied Biosystem), and 10 pmol of each forward and reverse primers. After an initial denaturation of 10 min at 94°C, 32 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s, with a final extension at 72°C for 2 min, were performed. After checking for amplification, the PCR products were gel purified using the Qiaquick gel extraction kit (Qiagen, Crawley, UK) and sequenced using the Applied Biosystem Big Dye Primer Chemistry and ABI3100 Prism® Sequencer (Applied Biosystems).
Sequence analysis of the four individuals identified a single-point mutation, G to A, altering amino-acid position 156 in exon 3, Ala (GCC) to Thr (ACC) (dbSNP ref rs3796130). The G to A mutation creates an Hph1 site. The PCR was used to specifically amplify a region of GPIX sequence encompassing codon 156 in all members of the Asian Indian family, and the amplicon restricted with Hph1 to screen for the Ala 156 (GCC) to Thr (ACC) mutation. A TaqMan assay was designed to type the G → A mutation in 300 unrelated North European subjects aged 18–79 years randomly selected from the Twins UK cohort (www.twinsuk.ac.uk) in a 10 μl reaction volume containing TaqMan Universal PCR Master Mix (ABI) and 20 ng genomic DNA. The samples were run and analyzed on an ABI 7900HT analyzer (Applied Biosystems).
The THPO gene was sequenced in the four individuals selected from the family. All six exons, and at least 100 bp of the flanking intronic sequence, 250 bp of 5′, and 200 bp of 3′ flanking sequence were specifically amplified by the PCR in seven partially overlapping fragments. Design of the PCR primers was based on the THPO gene sequence according to Ensembl gene ID ENST00000204615. PCR and sequencing were performed using conditions and approach exactly as described for the GPIX gene. Five SNPs were identified in the four individuals: T/C in exon 2 (5′UTR), dbSNP ref rs956732; G/A in intron 3 (dbSNP ref rs2280740); C/A in intron 5 (dbSNP ref rs6142); G/A in exon 6, 3′UTR (db SNP ref rs6141); and G/A in exon 6, 3′UTR (24 bp downstream of rs6141). The ‘A’ allele of the rs6141 SNP in the 3′ UTR creates a site for the restriction enzyme NlaIII, while the ‘G’ allele removes the site. The SNP in exon 6 downstream of rs6141 is novel, the ‘A’ allele creates a site for NlaIV, while the ‘G’ allele removes the NlaIV site in this region. All individuals of the Asian Indian kindred were screened for both the 3′UTR SNPs by NlaIII and NlaIV restriction analysis, respectively, of specifically amplified DNA encompassing exon 6 of the THPO gene. Sequences of all the primers used are available on request.
Statistical analyses
Single-point linkage analysis was carried out under a major QTL inheritance model using the ILINK routine of the program FASTLINK.22, 23, 24 The QTL has two alleles, A1 and A2. The major QTL inheritance model has five parameters: the frequency of the A1 allele, p; three genotype-specific trait means, μA1A1, μA1A2, and μA2A2; and the variance of the QTL genotype-specific distribution, σ2. The three QTL genotypes were assumed to have equal trait variance. Multipoint linkage analysis was carried out across 91 cM of chromosome 3q using the program Vitesse.25, 26 The data was analyzed under the same major QTL inheritance model that was used in the single-point analysis. Multipoint location scores were computed at intervals across the test region and converted to lod scores by dividing the score by 4.6. The effects of the covariates were estimated by linear regression. Platelet counts were adjusted for the covariates by multiplying the counts by the estimated regression parameter. Statistical analyses of individual polymorphisms, including analysis of variance and regression analysis, were carried out using the publicly available statistical software called R.
Results
The mean and standard deviation of the observed platelet count distribution were 284 700/μl and 75 800/μl, respectively, and the minimum and maximum counts were 83 000/μl and 653 000/μl, respectively. A log transformation of scale was applied to the platelet count data to improve the fit to the normal distribution. Platelets counts were significantly affected by age, sex and β thalassemia. The sizes of the covariate effects were estimated by linear regression and the trait values were adjusted for the measured effects. The three covariates combined were found to account for 21% of the phenotypic variance, with age, sex, and β thalassemia individually accounting for 13, 3, and 5% of the variation in the platelet data, respectively.
Platelet counts decreased with age, and tended to be higher in women and in β thalassemia heterozygotes. Platelet counts that were more than three standard deviations different from the sample mean were set to missing.
Genome-wide linkage analysis
The major QTL model parameters estimated from the kindred data were: p=0.22, μA1A1=−1.183, μA1A2=−0.748, μA2A2=0.473, and σ2=0.620. The parameter estimates suggested that a low platelet count phenotype was segregating in the kindred. Although the maximum likelihood estimates of the inheritance model parameters represent the best statistical fit of the platelet data to the major gene model, they are not necessarily the parameter values that will give the most power in a linkage analysis. The estimated model would not be powerful for linkage analysis, given that the within-genotype trait distributions overlap considerably and the QTL allele frequency is high. The inheritance model used in the genome-wide linkage analysis assumed that the QTL is associated with low trait levels, having the parameter values: p=0.01, μA1A1=−2.00, μA1A2=0.00, μA2A2=1.00, and σ2=1.00. Goring and Terwilliger27 have shown, for discrete traits, that an increase in power is achieved by using ‘strong’ linkage models, whereby the disease locus genotypes are assigned with high probability because of the rare mutation, high-penetrance model parameters. The increase in power comes at the expense of an inflated recombination fraction estimate. The recombination fraction parameter absorbs the error in the model that results from using the strong model parameters, whereas a weak model estimated from the data will be less informative, have less error, have a smaller recombination fraction estimate, and give a lower lod score. Given that the true model is never known with certainty in genetic studies of complex traits, the inheritance model will always be incorrect and the recombination fraction will always be biased. Therefore, the upward bias that comes from using a powerful model, which will give higher lod scores, is of minimal concern.
A simulation study was carried out to assess the null distribution of lod scores given the genetic model used in the genome-wide linkage analysis. Lod scores were computed for 10 000 replicate data sets of the observed platelet counts and an unlinked marker having five equally frequent alleles. There were 236 replicates that had lod scores between 1.0 and 2.0 (probability based on 10 000 replicates equals 0.024), and 11 replicates with lod scores between 2.0 and 3.0; no lod scores greater than 3.0 were computed from the simulated data. If all 226 markers in the genome-screen were independent (which they are not) an overconservative estimate is that one could expect five to six markers showing lod scores between 1.0 and 2.0 by chance alone.
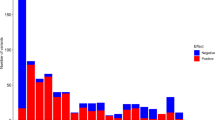
Figure 1 shows the single-point lod scores for the 226 markers tested across the genome. The greatest evidence for linkage was found on chromosome 3, given by both the value of the single-point lod score at an individual marker and the lod scores of adjacent markers. Table 1 shows the 11 markers that showed lod scores greater than 1.00 and recombination fraction estimates less than 0.50. Six consecutive markers across approximately 73 cM on chromosome 3 had lod scores greater than 1.00. The highest single-point lod score was 3.26 at marker D3S1309. Marker D16S413 on chromosome 16q24.3 showed a lod score of 1.96; the nearest marker was located 42.5 cM centromeric and did not show evidence for linkage. Markers D21S265 and D21S1270 (lod scores of 1.92 and 1.45, respectively) are separated by a genetic distance of 7 cM.
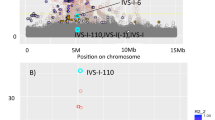
Multipoint analysis was carried out using a map of six markers spanning 91 cM of chromosome 3q (Figure 2). Locations are given as cM distances from the most centromeric marker D3S1271. A maximum multipoint lod score of 4.11 was observed at position 73.0 cM, the location of D3S1282. A one lod-unit confidence interval extends approximately 24 cM around the linkage peak between the 60 and 84 cM locations. A second peak in the multipoint curve occurs near the 45 cM position with a maximum lod score of 3.52 at the 41 cM position, the location of marker D3S1309. The one lod-unit confidence interval around this second peak extends approximately 27 cM between locations 28 and 55 cM. Positive lod scores across the six markers on chromosome 3q were observed along multiple lineages within the kindred. The largest family in the kindred contributed the highest lod scores under both peaks while the second smallest family in the kindred contributed the second highest lod score under the more telomeric peak; it does not share any founders with the larger family. The second largest family provided no evidence for linkage to chromosome 3q. The two peaks are the result of a dip in the multipoint lod score curve at the 56 cM location of the marker D3S1279. As expected, excluding marker D3S1279 results in one large multipoint peak across the region (results not shown). In order to determine if the dip in the multipoint curve was a result of an obligate recombination event imposed by the linkage analysis model, the allele frequency parameter was increased from 0.05 to 0.30 and the data were reanalyzed. The lod scores were considerably lower across the region and the same two-peak pattern was observed (results not shown).
Multipoint lod score curves across 91 cM of chromosome 3q. Approximate marker and gene locations are given on the lower and upper x-axes, respectively. For all three results, platelet count data were adjusted for the effects of age, sex, and β thalassemia. ‘GPIX adjusted’ indicates that platelet counts were adjusted for the effect of the Ala 156 to Thr mutation. ‘TPHO adjusted’ indicates that platelet counts were adjusted for the effect of the rs6141 (+24) G/A polymorphism in the 3′-UTR. ‘No adjustment’ indicates no adjustment for either of the genetic (1) variants. ‘Variance Comp’ indicates the analysis that was carried out using the model-free variance components approach implemented in the computer program SOLAR.
In order to verify the chromosome 3q results with an alternative linkage analysis method, multipoint linkage analysis using the model-free variance components approach implemented in the program SOLAR28 was carried out (Figure 2). The model-free approach shows a similar, although less significant, two-peak pattern across chromosome 3q, demonstrating the significant statistical power that was achieved with the strong model-based analysis of the kindred.
Candidate gene analysis of chromosome 3q
Within the linked region there were two genes that were known to be involved in the regulation of platelet count, thrombopoietin (THPO) and platelet glycoprotein IX (GPIX). GPIX and THPO map to chromosome 3q21.3 and 3q27.1, the respective centromeric and telomeric peaks in the multipoint curve. TPHO is a 353-amino-acid glycoprotein hormone produced primarily by the liver and is a major physiological regulator of circulating platelet count. TPHO affects the proliferation and differentiation of megakaryocytes, and, in general, circulating blood levels of TPHO are inversely related to blood platelet levels in human and animal models.5, 6, 7, 8 An activating splice donor mutation in the THPO gene has been shown to cause thrombocythaemia, a chronic myeloproliferative syndrome that results in elevated levels of circulating platelets.29 Five SNPs were found in the THPO gene in the kindred, including one previously unreported G/A variant in the 3′ untranslated region, and was genotyped in the full kindred.
GPIX is a relatively small glycoprotein of 177 amino acids found on the surface of human platelets. GPIX is a component of the platelet membrane glycoprotein Ib/IX/V complex, the platelet receptor for von Willebrand factor (vWF). Binding between the GP complex and vWF initiates platelet activation, and the complex is believed to regulate megakaryocytopoiesis, therefore affecting circulating platelet counts.30 Mutations in GPIX have been shown to cause BSS, a rare autosomal recessive disorder characterized by prolonged bleeding time, mild to severe thrombocytopenia, and giant platelets. A known nonsynonymous mutation at nucleotide position 653 was found resulting in an Ala 156 (GCC) to Thr (ACC) replacement. This mutation has recently been reported to cause BSS in a Chinese individual,18 although in this paper, the mutation was reported to occur at amino acid 140, which, in fact, is position 156 of the immature protein. The same mutation was previously found in a screen of 24 Japanese individuals for the purpose of SNP identification.31 The mutation was genotyped in the full kindred as well as 300 individuals of North European origin.
Analysis of variance was carried out to measure the effect of the GPIX mutation and the THPO 3′ UTR polymorphisms on platelet count variation. The analysis did not account for the correlation between observations due to familial relationships; therefore, the calculated P-values are overly liberal. Haplotypes were constructed for the two SNPs in the 3′ UTR of THPO. The novel THPO G/A polymorphism (24 bp downstream of rs6141) accounted for as much phenotypic variation as the haplotype, with the rs6141 SNP individually having less of an effect. The GPIX mutation and the THPO rs6141 (+24) site both showed statistically significant associations (P-value <0.05) with platelet counts, accounting for an estimated 6% (P-value=0.002) and 4% (P-value=0.016) of the variance in age-, sex-, and β-thalassemia-adjusted platelet counts, respectively, under a dominant model. The frequencies of the low trait associated alleles for the THPO rs6141 (+24) G/A polymorphism and the GPIX mutation were 0.06 and 0.09, respectively, including all typed individuals. When all carriers of the GPIX mutation were excluded from the analysis, 6% (P-value=0.007) of the phenotypic variation in platelets was accounted for by the THPO variant. Conversely, when all carriers of the minor THPO allele were excluded, the GPIX mutation accounted for 11% (P-value=0.0004) of platelet count variance in the kindred. A total of 300 individuals of North European descent were screened for the GPIX mutation to estimate its frequency. Only one copy of the allele was observed, suggesting that the mutation is rare in the European lineage.
Two new platelet count phenotypes were created by further adjusting the data for either the THPO rs6141 (+24) SNP or the GPIX mutation, and multipoint linkage analyses were carried out using the six markers across chromosome 3q (Figure 2). Removing the effects of the polymorphisms in GPIX or THPO results in large reduction in the lod scores across the chromosome 3q region. There are 11 individuals who are heterozygous at both genes so that adjusting for either polymorphic site will have a significant effect on the inheritance pattern of platelet counts at the other linked site, thereby reducing the lod score across the entire 3q region. There was one homozygote for the GPIX Ala156 Thr mutation in the kindred. The individual did not show symptoms of BSS or an abnormally low platelet count. He was heterozygous for β thalssaemia.
Three different haplotypes defined by the A allele at the GPIX mutation and the D3S1309 microsatellite (closest marker) were observed among six founders carrying the mutation in the kindred. THPO maps closest to marker D3S1282. Three haplotypes were also observed with the A allele at the THPO rs6141 (+24) polymorphism and the D3S1282 microsatellite in five founders carrying the THPO allele. All of the founder haplotypes segregate at least one generation in the kindred.
Discussion
Like most quantitative traits, there are rare Mendelian disorders that account for the extreme tails of the platelet count trait distribution; however, the genetic etiology underlying the majority of the trait distribution is complex and poorly understood. This study is the second genome-wide search for QTLs controlling platelet count that has been reported; the previous study showed no evidence for linkage to chromosome 3q17 but a suggestive linkage to 19q. We did not replicate the findings in the Australian study. However, the linkage results reported here are significant, with lod scores exceeding 3.0 in two regions of chromosome 3q. The results of the genetic analysis of chromosome 3q suggest that the two regions of significant linkage are the result of two QTL, as opposed to a drop in lod score due to the linkage information from a single marker in the region. There were two strong candidate genes for platelet count on chromosome 3q, platelet glycoprotein IX (GPIX) and thrombopoietin (THPO), and each gene mapped under one of the two peaks of statistical significance.
A rare functional mutation at nucleotide 2113 in GPIX results in an Ala 156 (GCC) to Thr (ACC) replacement and was shown to associated with low platelet counts in the kindred. The mutation creates a hydrophilic amino acid (Thr) from a hydrophobic amino acid (Ala) in the transmembrane domain of the mature protein. Wang et al18 recently reported the same mutation, although the amino-acid location cited was 140, in a Chinese woman with BSS. Using flow cytometric analysis, Wang et al18 showed that the patient's platelets had greatly reduced GPIbα and GPIX cell surface expression. Transient transfection studies by the same group confirmed that the mutation prevents GP Ib/IX complex insertion in the cytoplasmic membrane of platelets, and suppresses GPIbα and GPIX expression, but does not cause its intracellular degradation. The Chinese patient was homozygous for the mutation; however, heterozygous relatives did not show the characteristic BSS symptoms, leading the authors to conclude that the mutation acted under a recessive model of inheritance. In the current study, one individual was homozygous for the mutation in the Asian Indian kindred, but the platelet counts in this individual were comparable to those of the heterozygotes. The results of the two studies suggest that other unknown genetic factors exist which modify the penetrance of the GPIX transmembrane mutation, either compensating for the loss of function or causing the progression to BSS. Expression of normal GPIX is an important event in the development and differentiation of the megakaryocyte, culminating in the release of functional platelets. The morphologically abnormal platelets and MKs in BSS suggest that the normal expression of GPIX, and the GPIb/IX complex, is necessary for normal terminal differentiation of MKs.30 There is evidence that expression of the GPIb/IX complex plays a role in regulating megakaryopoiesis, first by inducing expression of the cyclin-dependent kinase inhibitor p21, and by inducing G1 cell cycle arrest.30 Nine other mutations in the GPIX gene have previously been identified in BSS patients; eight occur in the extracellular domain, with the other in the signal peptide of the protein.
A SNP in the 3′ UTR of the THPO gene was associated with platelet counts in the Asian Indian kindred. The TPHO glycoprotein hormone directly stimulates platelet production. The rs6141 (+24) G/A polymorphism appears to be rare, as it has not been previously reported despite extensive sequence analysis of the gene owing to its biological importance. It is possible that the 3′ UTR polymorphism is in near similar frequency and tightly linked to another QTL between chromosome 3q25.1 and 3q27.2. However, it is unlikely that a different genetic variant in TPHO is responsible for the linkage in the region. There is increasing evidence that 3′ UTRs of mRNAs contain regulatory elements that have important roles in post-transcriptional control of gene expression,32 but determining the role of the 3′ UTR polymorphism in THPO expression will require biological experiments.
The model-based approach used in the linkage analysis was powerful for detecting the QTL in the large kindred. The major QTL model parameters estimated from the data suggested that a locus for low platelet count was segregating in the kindred. The estimated inheritance model parameters were not used in the linkage analysis because they provided little QTL genotype information; the three possible QTL genotype probabilities for any one individual will all be similar, given the phenotype and genetic model parameters. This lack of strong locus genotype information results in a loss of power. Platelet count is a complex trait, influenced by multiple genetic and environmental factors, even within the single large kindred. Therefore, the expectation was that small groups of close relatives within the kindred would show simple inheritance patterns, while the overall pattern of inheritance would be complex. The model used in the analysis was designed to achieve the maximum lod score contribution from the small family groups that showed Mendelian inheritance patterns. Linkage analysis using the model-free, multipoint, variance component-based approach implemented in the program SOLAR28 also showed evidence for linkage to chromosome 3q. While the same two-peak pattern across chromosome 3q was observed using the model-free and model-based approaches, the strong model-based approach showed significantly higher lod scores.
A major advantage of large families in genetics studies is that they are very powerful for detecting linkage, particularly for rare mutations with strong effects on the phenotype. The large family design reduces the heterogeneity that plagues designs that rely on large numbers of small family units. The large number of meioses increases inheritance and linkage information and allows for the identification of marker mis-typings. On the other hand, a pitfall of studying one or a few large families is also demonstrated by this study. While one's chances of detecting significant linkage are increased with large families, the ability to identify the underlying gene and etiological mutation using the same resource is compromised. Linkage will extend over longer regions when detected in few families and lines between linkage and association become blurred. Hence, proving that a candidate allele is causative by genetic analysis of a single kindred is nearly impossible.
There were additional limitations to the study beyond those that are inherent in large pedigree studies. This study used an existing 20 cM resolution, genome-wide microsatellite marker data set that was generated in 1995. A 10 cM microsatellite marker map, as is the standard density used today, would have provided additional linkage information and more power to detect QTLs in the kindred. Platelet counts are affected by circadian fluctuations that were not taken into account in the collection of the phenotype data or in the statistical analysis and added an additional source of variation to the trait distribution. The association analysis was carried out using related individuals and will therefore give liberal P-values.
Two strong candidate genes for platelet count have been presented here. The argument in favor of their involvement in the trait comes from a combination of their location with respect to the linkage results, their known biological involvement in platelet count regulation, and the evidence in favor of the genetic variants being etiological. The evidence that the THPO 3′ UTR G/A polymorphism influences the protein's function is circumstantial; however, the biological evidence in favor of the gene itself is as strong as one can expect for a candidate gene. The Ala 156 to Thr mutation in GPIX has been shown to alter the protein's function; however, the link between mutated glycoprotein and lowered platelet counts is not completely clear. The two candidate QTLs appear to be rare, at least outside of the Asian Indian population. The results suggest that there may be genetic heterogeneity between Asian and European populations, which may explain the lack of consensus between the genome-scans. Genetic studies of individuals from additional populations, as well as independent samples from the Asian Indian population, will be needed to assess the role and frequency of these QTLs.
References
Eaton DL, de Sauvage FJ : Thrombopoietin: the primary regulator of megakaryocytopoiesis and thrombopoiesis. Exp Hematol 1997; 25: 1–7.
Kaushansky K : Thrombopoietin: understanding and manipulating platelet production. Annu Rev Med 1997; 48: 1–11.
Yang C, Li YC, Kuter DJ : The physiological response of thrombopoietin (c-Mpl ligand) to thrombocytopenia in the rat. Br J Haematol 1999; 105: 478–485.
Fielder PJ, Gurney AL, Stefanich E et al: Regulation of thrombopoietin levels by c-mpl-mediated binding to platelets. Blood 1996; 87: 2154–2161.
Emmons RV, Reid DM, Cohen RL et al: Human thrombopoietin levels are high when thrombocytopenia is due to megakaryocyte deficiency and low when due to increased platelet destruction. Blood 1996; 87: 4068–4071.
Wendling F : Thrombopoietin: its role from early hematopoiesis to platelet production. Haematologica 1999; 84: 158–166.
Kuefer MU, Wang WC, Head DR et al: Thrombopoietin level in young patients is related to megakaryocyte frequency and platelet count. J Pediatr Hematol Oncol 1998; 20: 36–43.
Tomita N, Motomura S, Sakai R et al: Strong inverse correlation between serum TPO level and platelet count in essential thrombocythemia. Am J Hematol 2000; 63: 131–135.
de Sauvage FJ, Hass PE, Spencer SD et al: Stimulation of megakaryocytopoiesis and thrombopoiesis by the c-Mpl ligand. Nature 1994; 369: 533–538.
Breton-Gorius J, Vainchenker W : Expression of platelet proteins during the in vitro and in vivo differentiation of megakaryocytes and morphological aspects of their maturation. Semin Hematol 1986; 23: 43–67.
Nurden P, Nurden A : Giant platelets, megakaryocytes and the expression of glycoprotein Ib-IX complexes. C R Acad Sci III 1996; 319: 717–726.
Hourdille P, Pico M, Jandrot-Perrus M, Lacaze D, Lozano M, Nurden AT : Studies on the megakaryocytes of a patient with the Bernard–Soulier syndrome. Br J Haematol 1990; 76: 521–530.
Feng S, Christodoulides N, Kroll MH : The glycoprotein Ib/IX complex regulates cell proliferation. Blood 1999; 93: 4256–4263.
Buckley MF, James JW, Brown DE et al: A novel approach to the assessment of variations in the human platelet count. Thromb Haemost 2000; 83: 480–484.
Evans DM, Frazer IH, Martin NG : Genetic and environmental causes of variation in basal levels of blood cells. Twin Res 1999; 2: 250–257.
Garner C, Tatu T, Reittie JE et al: Genetic influences on F cells and other hematologic variables: a twin heritability study. Blood 2000; 95: 342–346.
Evans DM, Zhu G, Duffy DL, Montgomery GW, Frazer IH, Martin NG : Multivariate QTL linkage analysis suggests a QTL for platelet count on chromosome 19q. Eur J Hum Genet 2004; 12: 835–842.
Wang Z, Zhao X, Duan W et al: A novel mutation in the transmembrane region of glyco-protein IX associated with Bernard–Soulier syndrome. Thromb Haemost 2004; 92: 606–613.
Thein SL, Sampietro M, Rohde K et al: Detection of a major gene for heterocellular hereditary persistence of fetal hemoglobin after accounting for genetic modifiers. Am J Hum Genet 1994; 54: 214–228.
Craig JE, Rochette J, Fisher CA et al: Dissecting the loci controlling fetal haemoglobin production on chromosomes 11p and 6q by the regressive approach. Nat Genet 1996; 12: 58–64.
Garner C, Mitchell J, Hatzis T, Reittie J, Farrall M, Thein SL : Haplotype mapping of a major quantitative-trait locus for fetal hemoglobin production, on chromosome 6q23. Am J Hum Genet 1998; 62: 1468–1474.
Cottingham Jr RW, Idury RM, Schaffer AA : Faster sequential genetic linkage computations. Am J Hum Genet 1993; 53: 252–263.
Schaffer AA : Faster linkage analysis computations for pedigrees with loops or unused alleles. Hum Hered 1996; 46: 226–235.
Idury RM, Elston RC : A faster and more general hidden Markov model algorithm for multipoint likelihood calculations. Hum Hered 1997; 47: 197–202.
O'Connell JR : Rapid multipoint linkage analysis via inheritance vectors in the Elston–Stewart algorithm. Hum Hered 2001; 51: 226–240.
O'Connell JR, Weeks DE : The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance [see comments]. Nat Genet 1995; 11: 402–408.
Goring HH, Terwilliger JD : Linkage analysis in the presence of errors IV: joint pseudomarker analysis of linkage and/or linkage disequilibrium on a mixture of pedigrees and singletons when the mode of inheritance cannot be accurately specified. Am J Hum Genet 2000; 66: 1310–1327.
Almasy L, Blangero J : Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 1998; 62: 1198–1211.
Wiestner A, Schlemper RJ, van der Maas AP, Skoda RC : An activating splice donor mutation in the thrombopoietin gene causes hereditary thrombocythaemia. Nat Genet 1998; 18: 49–52.
Eisbacher M, Khachigian LM, Khin TH, Holmes ML, Chong BH : Inducible expression of the megakaryocyte-specific gene glycoprotein IX is mediated through an Ets binding site and involves upstream activation of extracellular signal-regulated kinase. Cell Growth Differ 2001; 12: 435–445.
Haga H, Yamada R, Ohnishi Y, Nakamura Y, Tanaka T : Gene-based SNP discovery as part of the Japanese Millennium Genome Project: identification of 190,562 genetic variations in the human genome. Single-nucleotide polymorphism. J Hum Genet 2002; 47: 605–610.
Hesketh J : 3′-Untranslated regions are important in mRNA localization and translation: lessons from selenium and metallothionein. Biochem Soc Trans 2004; 32: 990–993.
Acknowledgements
This work is supported in parts by a grant (5 R01 HL69259-03) from the National Institutes of Health (NIH/NHLBI) to Swee Lay Thein and Chad Garner, and by a grant (G0000111) from the Medical Research Council UK to Swee Lay Thein. The Twins UK cohort is supported by the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Garner, C., Best, S., Menzel, S. et al. Two candidate genes for low platelet count identified in an Asian Indian kindred by genome-wide linkage analysis: glycoprotein IX and thrombopoietin. Eur J Hum Genet 14, 101–108 (2006). https://doi.org/10.1038/sj.ejhg.5201499
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201499
Keywords
This article is cited by
-
Genomic Variation Affecting MPV and PLT Count in Association with Development of Ischemic Stroke and Its Subtypes
Molecular Neurobiology (2023)
-
New gene functions in megakaryopoiesis and platelet formation
Nature (2011)
-
Genome-wide association study of hematological and biochemical traits in a Japanese population
Nature Genetics (2010)
-
Bernard-Soulier syndrome (Hemorrhagiparous thrombocytic dystrophy)
Orphanet Journal of Rare Diseases (2006)
-
A systematic analysis of disease-associated variants in the 3′ regulatory regions of human protein-coding genes II: the importance of mRNA secondary structure in assessing the functionality of 3′ UTR variants
Human Genetics (2006)