Abstract
Crigler–Najjar syndrome type I (CN-I) is a rare and severe autosomal recessive metabolic disease due to a total deficiency of bilirubin uridine diphosphate glucuronosyltransferase located on chromosome 2. We report on a child with CN-I due to a phenylalanine residue deletion inherited only from the father carrying this deletion at the heterozygous state. Cytogenetic analyses showed no deletion of the chromosomal 2q37 region. Microsatellite analysis of the child and his parents was consistent with paternal isodisomy for chromosome 2 in the child. This report demonstrates that uniparental disomy may be at the origin of very rare diseases transmitted as autosomal recessive traits and emphasizes the need for parental DNA analysis in such cases.
Similar content being viewed by others
Introduction
Uniparental disomy (UPD) is a rare cytogenetic phenomenon that occurs when both homologs of a chromosomal pair come from a single parent. Uniparental heterodisomy is defined by the presence of two different homologs, whereas uniparental isodisomy is defined by the presence of two identical homologs (same genetic information).1 Many mechanisms can explain UPD occurring during meiosis and/or mitosis: trisomic reduction to disomy, monosomic rescue, gamete complementation, mitotic errors (loss of one chromosome and duplication of the remaining chromosome).2 A survey of reported cases demonstrates a preponderance of trisomy rescue, which is frequently associated with confined-to-placenta mosaicism.3 Maternal UPD is more frequent than paternal UPD (approximately 3:1).3
UPD for chromosome 2 has been rarely reported.4, 5, 6, 7, 8, 9, 10 In most reports, maternal UPD for chromosome 2 with trisomy 2 in mosaic confined to placenta was associated with intrauterine growth retardation.4, 5, 6, 7 Maternal UPD, which is more often heterodisomy (probably the consequence of trisomy rescue), does not seem clinically relevant. In one case, reported by Spiekerkoetter, maternal UPD associated with isodisomy was responsible for trifunctional protein deficiency.8 Only two cases of paternal UPD for chromosome 2 have been reported. Unlike maternal UPD, all cases of paternal UPD were associated with isodisomy. Thompson et al9 reported that, in one patient, retinal dystrophy was due to paternal isodisomy with homoallelism for mutations in MERTK. The authors also confirmed that there are no paternally imprinted genes on chromosome 2 that have a major effect on phenotype. A second case of paternal UPD for chromosome 2 reported by Chavez et al10 has been responsible for steroid 5alpha-reductase 2 deficiency.
Crigler–Najjar syndrome type I (CN-I, MIM 218800) is a rare (less than 1/106 live births) and severe autosomal recessive disorder. CN-I is caused by the deficiency of uridine diphosphate glucuronosyltransferase (UGT1A1), the hepatic enzyme responsible for bilirubin elimination.11 The disease manifests itself during the first hours of life with severe and persistent unconjugated hyperbilirubinemia, often leading to exchange transfusion. Affected infants are at high risk for bilirubin-induced brain damage (kernicterus). Since the cloning of the gene encoding UGT (located on 2q37.1), about 30 different mutations have been reported,12, 13, 14 including several founder effects in countries such as Tunisia and Sardinia.15, 16, 17
We report here the case of a male infant with CN-I syndrome in whom molecular studies allowed us to recognise paternal isodisomy for chromosome 2.
Materials and methods
Case report
This male infant was the first child of healthy unrelated parents. The father was 26 years old and the mother 24. He was born at full-term after an uneventful pregnancy (weight 3350 g, height 51 cm). He became jaundiced in the hours following birth. Save jaundice and mild axial hypotonia, physical examination proved normal. At hour 24, serum bilirubin concentration was 350 μmol/l, entirely unconjugated, whereas red blood cell and white blood cell counts were normal. Baby's and mother's blood groups were A+, and Coombs test was negative. G6PD and pyruvate kinase activities were normal. The infant received continuous intensive phototherapy during 24 h, resulting in a decrease of bilirubin serum concentration to 280 μmol/l. During the following days, the infant had to stay between 10 and 12 h a day under phototherapy in order to keep the serum bilirubin concentration below 350 μmol/l. At 3 weeks, the diagnosis of CN-I was suspected and blood was sampled for molecular studies. The child was discharged at 3 months with at-home phototherapy, 10–12 h a day. His parents moved to another city and, a few weeks later, owing to extra-pyramidal manifestations, kernicterus was suspected and related to a long period of elevated unconjugated hyperbilirubinemia. The child was then regularly followed up and psychomotor delay became obvious (he was not able to sit alone at 11 months) with bouts of opisthotonos and extra-pyramidal hypertonia, while serum bilirubin concentration was between 320 and 360 μmol/l. At 1 year, the child was found dead in his bed. Permission for post-mortem examination was refused by his parents.
UGT1A1 gene analysis
Genomic DNA was extracted from peripheral leukocytes. The five exons of the UGT1A1 gene and the flanking intron–exon junctions were PCR amplified using previously described primers and experimental conditions.12 SSCP analysis was used to screen and the five exons were sequenced directly. TATA box genotyping was performed as reported previously.18
Microsatellites analysis
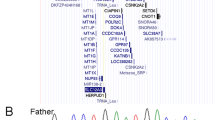
Genomic DNA was isolated from blood leukocytes. Polymorphic markers D2S423, D2S337, D2S113, D2S114, D2S1326, D2S2237 and D2S125 from chromosome 2 were PCR amplified (see location on chromosome 2 in Figure 1). Oligoprimer sequences were obtained from The Wellcome Trust Sanger Institute. Polymerase chain reactions were performed in a GeneAmp PCRsystem 9700 with denaturation for 2 min at 95°C, followed by 30 cycles of 40 s at 94°C, 30 s at 53 or 55°C and 1 or 2 min at 72°C with a final extension of 7 min at 72°C. DNA was amplified in a total volume of 50 μl containing 30 ng of DNA, 1 mM MgCl2, 300 μ M DNTPs and 0.4 μ M forward and reverse primers.
The PCR product was mixed with loading buffer in 1:1 proportion and loaded onto an 8% polyacrylamide 29:1 sequencing gel. Electrophoresis was performed on a Sequi-Gen Sequencing Cell (BioRad, Hercules, CA, USA). Bands were visualized by UV light after ethidium bromide coloration.
Cytogenetic analysis
Chromosome analyses were performed on cultured lymphocytes using R banding techniques. Fluorescence in situ hybridization (FISH) was performed as described previously.19 Specific probes for 2q37 chromosomal region were used.
Results
In the proband, a homozygote trinucleotide deletion was identified in exon 1, thus resulting, in the protein, in the deletion of one of the two adjacent phenylalanine residues at positions 170 and 171. In the promoter of the UGT1A1 gene, the child was homozygous for the wild-type allele A(TA)6TAA. The father was heterozygous for the deletion and homozygous for the A(TA)6TAA normal allele in the promoter. Surprisingly, in the mother, no mutation was found in the five exons, including the exon–intron junctions, whereas she was homozygous for the A(TA)7TAA mutant allele in the promoter. These results were confirmed on two independent sequencing procedures.
The genetic analysis of the seven microsatellites revealed that five were informative for paternal disomy, of which three were informative for paternal isodisomy. Two microsatellites were noninformative (Figure 1). The infant karyotype was normal 46,XY. No trisomy 2 mosaicism was found in 300 cells analyzed. FISH using probes specific for 2q37 band (RP11-69j7, RP11-118M12, TelVysion 2q) showed no deletion (Figure 2).
Discussion
We report on an infant with CN-I in whom UPD was identified during familial molecular study. Crigler–Najjar was due to the deletion of one of the two phenylalanine residues at position 170 or 171. Exon 1 of wild-type UGT1A1 contains two consecutive codons (TTC TTC) encoding two phenylalanine residues at positions 170 and 171. Therefore, the deletion of Phe170 is indistinguishable from that of Phe171, on the basis of nucleotide or amino-acid sequence (Table 1). Deletion of one of these two TTC triplets has been reported previously to cause CN-I.20, 21 Experimental substitution of Phe170 abolished UGT1A1 activity, but that of Phe171 did not, suggesting that, in the wild-type enzyme, Phe170 is irreplaceable while the neighboring Phe171 is not. These two residues are located in a ‘membrane-embedded helix’ of UGT1A1. The modification of this hydrophobic region may have a particular impact on enzyme activity and could be responsible for the phenotype.22
UPD for chromosome 2 is a rare event,4, 5, 6, 7, 8, 9, 10 and only two cases of paternal UPD for chromosome 2 have been described.9, 10 Many mechanisms can explain UPD, but it seems that, for chromosome 2, trisomic zygote reduction to disomy is likely.4, 5, 6, 7, 8, 23, 24 Placental mosaicism for chromosome 2 has been reported previously,23 even though this is not the most frequent. As pointed out in this report, the pattern of aneuploid cells for each trisomy can be used to predict the relative contribution of meiotic and mitotic errors to confined placental mosaicism, and thus the incidence of UPD. Moreover, confined placental mosaicism for trisomy 2 shows evidence of a nonrandom distribution of aneuploid cells between the different extra-embryonic cell lineages.22 In our case, the mechanism of UPD is not identified clearly. However, the infant was born after an uneventful pregnancy and measurements at birth were normal (no intrauterine growth retardation, birth height and weight normal) contrary to cases with confined placental mosaicism for trisomy 2.4, 5, 6, 7, 8 No trisomy 2 mosaicism was found in analyzed blood cells. In this way, monosomy rescue, which is less likely associated with clinically relevant mosaicism, is the most likely mechanism to explain paternal isodisomy for chromosome 2.
CN-I is a very rare and severe disease transmitted as an autosomal recessive trait. Molecular studies have shown that it was a heterogeneous disease, more than 30 mutations having been identified (for a review, see Kadakol et al25). To date, in all cases, parental DNA analysis had shown that both parents were heterozygous for one mutation. This has also been used for performing prenatal diagnosis of CN-I.26 However, as pointed out by Spiekerkoetter,8 as UPD may be more frequent in very autosomal recessive diseases, it is mandatory to confirm mutations in parents before performing prenatal diagnosis. Moreover, the identification of UPD has a great impact on genetic counselling, reducing dramatically the 25% risk normally used for such diseases. However, in our case, genetic counselling should be careful. The A(TA)7TAA mutation paired with deleterious mutation could be responsible for CN-I or CN-II.27 Owing to parental genotypes, future children will be compound heterozygotes for the A(TA)7TAA mutant allele and the phenylalanine deletion. This genotype has never been reported but could be at the origin of neonatal prolonged and important unconjugated hyperbilirubinemia.
In conclusion, this report demonstrates that UPD may be at the origin of very rare diseases transmitted as autosomal recessive traits and emphasizes the need for parental DNA analysis in such cases.
References
Engel E : A new genetic concept: uniparental disomy and its potential effect, isodisomy. Am J Med Genet 1980; 6: 137–143.
Engel E, DeLozier-Blanchet CD : Uniparental disomy, isodisomy, and imprinting: probable effects in man and strategies for their detection. Am J Med Genet 1991; 40: 432–439.
Kotzot D : Abnormal phenotypes in uniparental disomy (UPD): fundamental aspects and a critical review with bibliography of UPD other than 15. Am J Med Genet 1999; 82: 265–274.
Hansen WF, Bernard LE, Langlois S et al: Maternal uniparental disomy of chromosome 2 and confined placental mosaicism for trisomy 2 in a fetus with intrauterine growth restriction, hypospadias, and oligohydramnios. Prenat Diagn 1997; 17: 443–450.
Wolstenholme J, White I, Sturgiss S, Carter J, Plant N, Goodship JA : Maternal uniparental heterodisomy for chromosome 2: detection through ‘atypical’ maternal AFP/hCG levels, with an update on previous case. Prenat Diagn 2001; 21: 813–817.
Harrison K, Eisenger K, Anyane-Yeboa K, Brown S : Maternal uniparental disomy of chromosome 2 in a baby with trisomy 2 mosaicism in amniotic fluid culture. Am J Med Genet 1995; 58: 147–151.
Webb AL, Sturgiss S, Warwicker P, Robson SC, Goodship JA, Wolstenholme J : Maternal uniparental disomy for chromosome 2 in association with confined placental mosaicism for trisomy 2 and severe intrauterine growth retardation. Prenat Diagn 1996; 16: 958–962.
Spiekerkoetter U, Eeds A, Yue Z, Haines J, Strauss AW, Summar M : Uniparental disomy of chromosome 2 resulting in lethal trifunctional protein deficiency due to homozygous alpha-subunit mutations. Hum Mut 2002; 20: 447–451.
Thompson DA, McHenry CL, Li Y et al: Retinal dystrophy due to paternal isodisomy for chromosome 1 or chromosome 2, with homoallelism for mutations in RPE65 or MERTK, respectively. Am J Hum Genet 2002; 70: 224–229.
Chavez B, Valdez E, Vilchis F : Uniparental disomy in steroid 5alpha-reductase 2 deficiency. J Clin Endocrinol Metab 2000; 85: 3147–3150.
Arias IM, Gartner LM, Cohen M, Ezzer JB, Levi AJ : Chronic nonhemolytic unconjugated hyperbilirubinemia with glucuronyl transferase deficiency. Clinical, biochemical, pharmacologic and genetic evidence for heterogeneity. Am J Med 1969; 47: 395–409.
Bosma PJ, Chowdhury NR, Goldhoorn BG et al: Sequence of exons and the flanking regions of human bilirubin-UDP-glucuronosyltransferase gene complex and identification of a genetic mutation in a patient with Crigler–Najjar syndrome, type I. Hepatology 1992; 15: 941–947.
Kadakol A, Ghosh SS, Sappal BS, Sharma G, Chowdhury JR, Chowdhury NR : Genetic lesions of bilirubin uridine-diphosphoglucuronate glucuronosyltransferase (UGT1A1) causing Crigler–Najjar and Gilbert syndromes: correlation of genotype to phenotype. Hum Mutat 2000; 16: 297–306.
Ritter JK, Yeatman MT, Ferreira P, Owens IS : Identification of a genetic alteration in the code for bilirubin UDP-glucuronosyltransferase in the UGT1 gene complex of a Crigler–Najjar type I patient. J Clin Invest 1992; 90: 150–155.
Labrune P, Myara A, Hadchouel M et al: Genetic heterogeneity of Crigler–Najjar syndrome type I: a study of 14 cases. Hum Genet 1994; 94: 693–697.
Rosatelli MC, Meloni A, Faa V et al: Molecular analysis of patients of Sardinian descent with Crigler–Najjar syndrome type I. J Med Genet 1997; 34: 122–125.
Francoual J, Rivierre A, Mokrani C et al: Crigler–Najjar syndrome type I in Tunisia may be associated with founder effect related to the Q357R mutation within the UGT1 gene. Hum Mutat 2002; 19: 570–571.
Le Bihan-Lefauvre B, Francoual J, Labrune P, Chalas J, Capel L, Lindenbaum A : Mise au point et intérêt du diagnostic de la maladie de Gilbert par biologie moléculaire. Ann Biol Clin 2001; 59: 61–66.
Aboura A, Coulomb-L'Hermine A, Audibert F, Capron F, Frydman R, Tachdjian G : De novo interstitial direct duplication 1(q23.1q31.1) in a fetus with Pierre Robin sequence and camptodactyly. Am J Med Genet 2002; 108: 153–159.
Ritter JK, Yeatman MT, Kaiser C, Gridelli B, Owens IS : A phenylalanine codon deletion at the UGT1 gene complex locus of a Crigler–Najjar type I patient generates a pH-sensitive bilirubin UDP-glucuronosyltransferase. J Biol Chem 1993; 268: 23573–23579.
Rosatelli MC, Meloni A, Faà V et al: Molecular analysis of patients of sardanian descent with Crigler–Najjar syndrome type I. J Med Genet 1997; 34: 122–125.
Ciotti M, Cho JW, George J, Owens IS : Required buried alpha-helical structure in the bilirubin UDP-glucuronosyltransferase, UGT1A1, contains a nonreplaceable phenylalanine. Biochemistry 1998; 37: 11018–11025.
Wolstenholme J : Confined placental mosaicism for trisomies 2, 3, 7, 8, 9, 16, and 22: their incidence, likely origins, and mechanisms for cell lineage compartmentalization. Prenat Diagn 1996; 16: 511–524.
Gibbons B, Cheng HH, Yoong AK, Brown S : Confined placental mosaicism for trisomy 2 with intrauterine growth retardation and severe oligohydramnios in the absence of uniparental disomy in the fetus. Prenat Diagn 1997; 17: 689–690.
Kadakol A, Sappal BS, Ghosh SS et al: Interaction of coding region mutations and the Gilbert-type promoter abnormality of the UGT1A1 gene causes moderate degrees of unconjugated hyperbilirubinaemia and may lead to neonatal kernicterus. J Med Genet 2001; 38: 244–249.
Francoual J, Trioche P, Mokrani C et al: Prenatal diagnosis of Crigler–Najjar syndrome type I by single-strand conformation polymorphism (SSCP). Prenat Diagn 2002; 22: 914–916.
Ciotti M, Chen F, Rubaltelli FF, Owens IS : Coding defect and a TATA box mutation at the bilirubin UDP-glucuronosyltransferase gene cause Crigler–Najjar type I disease. Biochim Biophys Acta 1998; 1407: 40–50.
Acknowledgements
We thank Dr Sophie Brisset for cytogenetical analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Petit, F., Gajdos, V., Parisot, F. et al. Paternal isodisomy for chromosome 2 as the cause of Crigler–Najjar type I syndrome. Eur J Hum Genet 13, 278–282 (2005). https://doi.org/10.1038/sj.ejhg.5201342
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201342
Keywords
This article is cited by
-
A compendium and functional characterization of mammalian genes involved in adaptation to Arctic or Antarctic environments
BMC Genetics (2017)
-
Maternal uniparental heterodisomy with partial isodisomy of a chromosome 2 carrying a splice acceptor site mutation (IVS9–2A>T) in ALS2 causes infantile-onset ascending spastic paralysis (IAHSP)
neurogenetics (2009)
-
Deletions of 2q14 that include the homeobox engrailed 1 (EN1) transcription factor are compatible with a normal phenotype
European Journal of Human Genetics (2006)
-
A fascination with chromosome rescue in uniparental disomy: Mendelian recessive outlaws and imprinting copyrights infringements
European Journal of Human Genetics (2006)