Abstract
Chronic mucocutaneous candidiases (CMC) are a group of rare disorders where an altered immune response against Candida leads to persistent and/or recurrent infections of the skin, nails, and mucous membranes. We analysed a five-generation Italian family with an isolated form of CMC, affecting nails only, in the presence of low serum concentration of intercellular adhesion molecule I (ICAM-1). We excluded linkage to candidate regions on chromosomes 2p (CMC with thyroid disease), 21q22.3 (APECED), and 19q13 (ICAM-1). We then carried out a genome-wide scan and assigned the CMC locus to a 19 cM pericentromeric region on chromosome 11.
Similar content being viewed by others
Introduction
Chronic mucocutaneous candidiases (CMC) are rare disorders where an altered immune response against Candida (mainly Candida albicans) leads to chronic infections of the skin, nails, and mucous membranes. Most CMC types present an early age of onset and are associated with altered phagocytosis and chemotaxis.1 To date, only two hereditary forms of CMC have been localised. The classical CMC (autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy, APECED; MIM *240300) caused by autosomal recessive mutations of the autoimmune regulator gene (AIRE) was mapped to chromosome 21q22.3,2,3 while the autosomal dominant CMC, associated with thyroid disease, was assigned to chromosome 2p.1
We previously reported a five-generation Italian family originating from a small village of Sicily, in which a distinct form of isolated familial chronic nail candidiasis is characterised by early onset infections restricted to the nails of the hands and feet, caused by different species of Candida, and associated with low serum concentration of intercellular adhesion molecule I (ICAM-1).4 Here we describe the exclusion of linkage to 21q22.3 (APECED), 2p, and 19p13 (ICAM-1) loci. We also report a genome-wide scan that mapped the disease locus to a 19 cM pericentomeric region on chromosome 11.
Patients and methods
Subjects
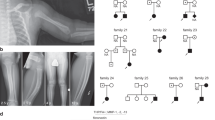
The family originates from a rural village in Sicily and includes 11 affected subjects in seven generations. A complete clinical description of the family has been reported.4 Briefly, all affected family members presented nail dystrophy with hyperkeratosis and dark and thick nails (Figure 1). They were affected by onycomycosis caused by Candida involving all the nails of hands and feet. Caustication was followed by regeneration of the nails, manifesting similar dystrophic features and Candida infection. The first symptoms of generalised onycomycosis occurred at the age of 3 months. All patients had been treated over the years by topical and systemic antimycotic drugs, which resulted in temporary recovery, while never reaching complete remission. All subjects underwent exhaustive tests excluding possible endocrine abnormalities. After informed consent, a blood sample was collected from seven affected and 15 unaffected family members.
Genotyping
Genome scanning was performed using 358 microsatellite markers at a distance of ∼10 cM (ABI PRISM™ Linkage Mapping Set Version 2). PCR was performed according to the manufacturer's protocol. To refine the region of linkage, additional markers were chosen from the Généthon map and all were labelled with the 6-FAM fluorochrome. PCR products were analysed on a model 3100 automated fluorescent DNA sequencer (Applied Biosystems). PCR reaction (1 μl) was combined with 10 μl of formamide and 0.5 μl of a fluorescent size marker (ROX 400HD). Each sample was run for 30 min. The fluorescent data collected during the run were analysed by the Genescan Analysis program (version 3.5). Each marker was examined by the Genotyper program (version 3.7), in order to analyse inheritance patterns and prepare the allele labels for export to linkage applications.
Linkage analysis
Linkage analysis was performed using the SimWalk package (version 2.82).5 SimWalk2 uses algorithms that can compute linkage statistics on highly complex pedigrees taking advantage of the power provided by the full-pedigree structure. Based on the presence of seven parent-to-child transmission, we carried out linkage analyses assuming dominant inheritance with reduced (0.90) penetrance. As the inbred structure of the family4 does not allow to rule out other modes of inheritance, we also used SimWalk2 Markov chain Monte Carlo (MCMC) methods to compute allele-sharing statistics and provide a model-free (or nonparametric) analysis. Files for SimWalk2 analysis were generated using Mega2 package (version 2.3 R2).6 Most-likely haplotypes were also calculated by SimWalk2. The order of the markers loci and the recombination distances used for linkage analysis were based on the Généthon linkage map.
Results
We first examined the known CMC loci on chromosomes 2p and 21q22.3 (APECED) and the ICAM-I region on chromosome 19q13, based on the low ICAM-I serum levels in the patients. In all three cases the negative LOD score values obtained allowed us to exclude an involvement of these regions in the investigated form of CMC.
A genome-wide screen was then conducted with microsatellite markers spaced at intervals of about 10 cM, and a multipoint LOD score of 3.46 was obtained for the pericentromeric region of chromosome 11 at marker D11S905. The haplotype analysis allowed to identify an initial 26 cM disease locus. Therefore, this region was refined using eight additional markers. A new maximum multipoint LOD score of 3.52 was achieved for the 19 cM region between D11S1312 and D11S4191 (Figure 2 ). Nonparametric analysis confirmed this location, as markers on chromosome 11 provided, by far, the strongest evidence for linkage in the genome (Table 1). Haplotype analysis disclosed a recombination event in the affected subject III:5, between markers D11S1312 and D11S4200, defining the upper limit of the disease interval. The lower limit was established by a recombination between markers D11S1350 and D11S4191 in the affected subject IV:3 (Figure 3).
Haplotypes of 11 DNA markers on chromosome 11p12–q12.1 in the examined branch of the family. The blackened bars indicate the haploidentical region in affected individuals that define the critical familial chronic nail candidiasis region between flanking markers D11S1312 and D11S4191. The arrows indicate the recombinations that define the critical region.
Discussion
We analysed a seven-generation Italian family with an isolated form of familiar chronic nail candidiasis. We excluded linkage to candidate regions on chromosomes 2p, and 21q22.3, showing that isolated familiar chronic nail candidiasis is not allelic to the form associated with thyroid disease or to APECED. We also investigated and excluded an involvement of the chromosome 19q13 region harbouring the ICAM-1 gene.
Both autosomal dominant and autosomal recessive inheritance models of CMC have been reported.7,8,9,10 The pedigree analysis of the examined family revealed the presence of seven parent-to-child transmissions that strongly favoured a dominant model over a recessive one. The presence of extensive inbreeding within the family was consistent with an autosomal recessive mode of inheritance. However, we could not identify a common ancestor who might have transmitted a recessive allele to all parents of the affected individuals. Given the rarity of the phenotype, the possibility of unrelated founders seemed unlikely, leading us to exclude pseudodominant inhertitance (i.e. recessive transmission mimicking dominance, owing to the presence of extensive inbreeding). We then conducted linkage analysis assuming autosomal dominant inheritance with reduced penetrance, as such a mode of inheritance was consistent with the hypothesis of a single founder, thereby offering a more parsimonious explanation of the observed pattern of disease transmission. We also sought a confirmation of locus assignment by model-free (nonparametric) methods. Haplotype and multipoint LOD score analysis conducted on the whole genome placed the familial chronic nail candidiasis locus within a 19 cM pericentromeric region on chromosome 11. Markers in this region also generated the maximum values for the SimWalk allele-sharing statistic and the maximum nonparametric LOD (NPL) scores of the entire genome (Table 1).
Examination of the region between D11S1312 and D11S4191 in the NCBI databases identified the catalase (CAT) gene, as a putative positional candidate. Interestingly, it has been demonstrated that the addition of catalase to culture medium reduces the killing of C. albicans, suggesting that the candidacidal activity is dependent on the myeloperoxidase system.11 Thus, one can speculate that the activating CAT mutations might cause CMC. This possibility is currently investigated. On the other hand, the candidate interval contains numerous transcripts and predicted genes. Therefore, the genetic investigation of additional families is mandatory to refine the disease locus and understand the mechanisms underlying this CMC disorder.
References
Atkinson TP, Schaffer AA, Grimbacher B et al: An immune defect causing dominant chronic mucocutaneous candidiasis and thyroid disease maps to chromosome 2p in a single family. Am J Hum Genet 2001; 69: 791–803.
Ahonen P: Autoimmune polyendocrinopathy–candidosis–ectodermal dystrophy (APECED): autosomal recessive inheritance. Clin Genet 1985; 27: 535–542.
Nagamine K, Peterson P, Scott HS et al: Positional cloning of the APECED gene. Nat Genet 1997; 17: 393–398.
Zuccarello D, Salpietro DC, Gangemi S et al: Familial chronic nail candidiasis with ICAM-1 deficiency: a new form of chronic mucocutaneous candidiasis. J Med Genet 2002; 39: 671–675.
Sobel E, Lange K: Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 1996; 58: 1323–1337.
Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP, Weeks DE: Mega2, a data-handling program for facilitating genetic linkage and association analyses. Am J Hum Genet 1999; 65: A436.
Sams Jr WM, Jorizzo JL, Snyderman R et al: Chronic mucocutaneous candidiasis. Immunologic studies of three generations of a single family. Am J Med 1979; 67: 948–959.
Jorizzo JL, Sams Jr WM, Jegasothy BV, Olansky AJ: Cimetidine as an immunomodulator: chronic mucocutaneous candidiasis as a model. Ann Intern Med 1980; 92: 192–195.
Wells RS, Higgs JM, Macdonald A, Valdimarsson H, Holt PJ: Familial chronic muco-cutaneous candidiasis. J Med Genet 1972; 9: 302–310.
Germain M, Gourdeau M, Hebert J: Case report: familial chronic mucocutaneous candidiasis complicated by deep candida infection. Am J Med Sci 1994; 307: 282–283.
Loyola W, Gaziri DA, Gaziri LC, Felipe I: Concanavalin A enhances phagocytosis and killing of Candida albicans by mice peritoneal neutrophils and macrophages. FEMS Immunol Med Microbiol 2002; 33: 201–208.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mangino, M., Salpietro, D., Zuccarello, D. et al. A gene for familial isolated chronic nail candidiasis maps to chromosome 11p12–q12.1. Eur J Hum Genet 11, 433–436 (2003). https://doi.org/10.1038/sj.ejhg.5200985
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200985
Keywords
This article is cited by
-
Genetic Susceptibility to Fungal Infections in Humans
Current Fungal Infection Reports (2012)