Abstract
Branchio-Oto-Renal (BOR) syndrome is transmitted as an autosomal dominant disorder, affects an estimated 2% of profoundly deaf children, and is caused by mutations in the human EYA1 gene. However, in up to half of the reported cases, EYA1 mutation screening is negative. This finding has been taken as evidence of genetic heterogeneity. Mutation screening of the coding region of EYA1 in a panel of families linked to chromosome 8 was conducted using SSCP and direct sequencing. Only one point mutation in five probands was detected. However, complex rearrangements, such as inversions or large deletions, were discovered in the other four patients using Southern blot analysis. These data suggest that more complex rearrangements may remain undetected in EYA1 since SSCP and sequencing were commonly used to detect mutations in this gene.
Similar content being viewed by others
Introduction
Branchio-Oto-Renal (BOR) syndrome (MIM 113650) is an autosomal dominant disorder first described by Melnick et al.1,2 and further delineated by Fraser et al.3 The prevalence is estimated at 1 : 40 000 in the general population, and the syndrome has been reported to occur in about 2% of profoundly deaf children.4 Clinical expression is highly variable within and among families, but typical manifestations are branchial arch anomalies (preauricular pits, branchial fistulae and pinnae abnormalities), hearing loss (conductive, sensorineural or mixed) and renal hypoplasia.5,6
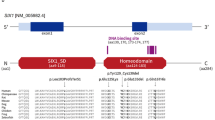
BOR syndrome was localised to chromosome 8q13.3 by linkage analysis7,8,9,10,11,12,1314 and deletion mapping.11,1415 Recently, a second locus was identified at chromosome 1q31.16 Mutations in EYA1 at 8q13.3, the human homologue of the Drosophila eyes absent gene, have been shown to underlie BOR syndrome17,18,19,20,2122 and Branchio-Oto (BO) syndrome in some families.22,23 The EYA1 gene is composed of seventeen exons spanning 156 kb and encodes a 559 amino acid protein. Exons 9 to 16 encode a highly conserved 271 amino acid domain, the eya homologous region (eyaHR), where most of the BO/BOR mutations have been found. Expression studies of EYA1 in mouse suggest a role in the development of the ear and kidney; consistent with these findings, EYA1 is highly expressed in foetal kidney and brain.17 In Drosophila, EYA1 is essential for eye formation24 and in humans, EYA1 mutations have been found in three patients with congenital cataracts.25
Since EYA1 was identified as the candidate gene for BOR, only about half of patients with the clinical diagnosis of either BO or BOR syndrome have been reported to carry mutations in EYA1.17,18,1920 Screening of the coding region of EYA1 in our panel of families by single-stranded conformation polymorphism (SSCP) analysis and direct sequencing detected only one alteration in five probands even though three families linked to the critical BOR region and two showed haplotype segregation consistent with linkage. However, we were able to detect deletions and complex rearrangements in the remaining four families using Southern blot analysis. Although some investigators have hypothesised that mutations in another gene in the same region could cause a BOR syndrome phenotype,26 our data suggest that the lack of confirmed mutations in EYA1 reflects a failure to screen for complex disease-causing genomic rearrangements.
Materials and Methods
Patients and concordance studies
Clinical details, pedigrees and linkage analysis of families K6015, K6030 and K6380 have been described previously.8,9
K6190 is a five-generation family reported to have typical but variable BOR features. Of 40 individuals, 20 affected persons have a variety of symptoms including hearing loss, branchial clefts or cysts and pre-auricular pits. Only one person has kidney failure. Some members of the family have learning problems, which may be related to the hearing loss. Peripheral blood samples were obtained from three affected individuals – the proband, her mother and her brother. The proband's father and grandfather had prelingual deafness, but no other symptoms characteristic of the BOR syndrome. Due to the lack of DNA samples on all members of K6190, concordance analysis was utilised to determine if the family should be included in the EYA1 mutation screening. A haplotype was constructed using 10 markers between D8S1060 and D8S84 to determine whether the proband, her brother and their affected mother shared a common haplotype flanking the EYA1 gene. Since the analysis did not exclude allelic association with the BOR phenotype, we included the K6190 family in the study.
K6310 is a three-generation family with five affected members sharing the phenotypic features of hearing loss, lop ear deformity, branchial fistula and kidney problems. Segregation of a presumed affected haplotype was consistent with linkage to the EYA1 region.
Cell lines and DNA samples
Genomic DNA was isolated from peripheral blood by high salt precipitation.27 Purified DNA was diluted to a concentration of 105 ng/μl. A lymphoblastoid cell line was established on the proband from K6190 using standard procedures.28
Analysis of single-strand conformation polymorphism (SSCP)
Primer sequences of the 17 exons of the human gene EYA1 were taken from Abdelhak et al.17 Ten μM of 5′OH-primers were end-labelled with 1 μCi at 3000 Ci/mmol [γ33P]-ATP (Andoteck, Irvine, CA, USA) and one unit of polynucleotide kinase (Promega, Madison, WI, USA) in standard buffer for 30 min at 37°C. Individual exons along with flanking intronic sequence were amplified using 50 ng of genomic DNA in a total volume of 50 μl containing 20 μM of dNTPs, 5 μM of each primer, and 0.056 μM of Taq polymerase (Sigma, St. Louis, MO, USA) with 0.02 μM of TaqStart™ Antibody (Clontech, Palo Alto, CA, USA). DNA was amplified for 35 cycles consisting of 1 min at 95°C, 1 min at 55°C, 1 min at 72°C, except for exons 2 and 9 where the annealing temperature used was 54°C. The last extension was lengthened to 5 min. One μl of the radiolabelled PCR product was denatured with 10 μl of stop solution (95% formamide, 10 mM NaOH, 0.25% Bromophenol Blue, 0.25% Xylene Cyanol) at 94°C for 2 min before cooling on ice. Three μl of the reaction mixture was loaded on a MDE™ Acrylamide gel (FMC, Rockland, ME, USA) prepared following the manufacturer's procedure. The migration was performed at a constant power of 7 watts for 14 h at room temperature. The gel was transferred onto Whatman chromatography paper (3MM Chr), and dried before exposure to X-ray Biomax film, (Kodak, Rochester, NY) using standard techniques.
Sequencing
PCR products were purified using Centricon® 100 concentrators (Amicon, Beverly, MA, USA), and directly sequenced using the Dye Terminator Cycle Sequencing Kit (PE Biosystems, Foster City, CA, USA). A final purification was performed on Centrisep columns (Princeton Separation, Adelphia, NJ, USA) before running the samples on an ABI Prism™ 373 sequencer (PE Biosystems). Exon 9 PCR products were subcloned into PCR2.1® (Invitrogen, CA, USA) following the manufacturer's procedure, prior to sequencing using M13 universal primer.
DNA hybridisation probes
Three cDNA clones covering overlapping regions of the EYA1 coding sequence were kindly provided by Dr C Petit (Institut Pasteur, Paris). The EYA1.1 clone contains the 5′UTR to exon 3 including exon 1′, the EYA1.2 clone contained exons 1 through 12, and the EYA1.3 clone contained exon 13 to the 3′UTR. The 3 EYA1 cDNA clones were digested with EcoRI and the plasmid inserts were gel-purified using QIAquick gel extraction kit (QIAGEN, Valencia, CA, USA) and labelled with 50 μCi at 3000 Ci/mmol [α32P]-CTP (NEN, Boston, MA, USA), using the random prime labelling system Redprime™ II (Amersham Pharmacia Biotech, Piscataway, NJ, USA). Additional cDNA probes, containing only exons 4 through 8 (EYA1.4), and exons 9 through 11 (EYA1.5), were generated by PCR amplification of the EYA1.2 clone (Table 1). Hybridisation of individual exons was performed using PCR products generated using the primer sets designed for SSCP analysis.17 Intronic probes were developed using primers residing in unique sequence found with the sequences of BAC clones RP11–11K9 (GenBank accession no. AC016465) and RP11–326E22 (GenBank accession no. AC022858) (Table 1). Each intronic probe was amplified using 100 ng of genomic DNA in a total volume of 50 μl, containing 20 μM of dNTPs, 10 μM of each primer and 0.056 μM of Taq polymerase. DNA was amplified for 35 cycles consisting of 1 min at 95°C, 30 s at appropriate annealing temperature and 1–2 min at 72°C. The last extension was lengthened from 5 to 7 min. Amplification was verified by agarose electrophoresis before gel-purification with the QIAquick gel extraction kit. Intronic probes were preassociated with sonicated human placental DNA (Sigma, St Louis, MO, USA) in 50 000X excess, at 65°C for 1.5 h.
Southern analysis
Five μg of each patient's genomic DNA were digested overnight at 37°C using one of the following enzymes: MspI, PstI, DraI, Sau3AI, HindIII, BamHI, PvuII (New England Biolabs®, Inc., Beverly, MA, USA). Digested DNA was electrophoresed on a 0.8% Seakem® LE agarose gel (BMA, Rockland, ME, USA) and transferred by alkali blotting to a Hybon-N+ membrane (Amersham Pharmacia Biotech, Piscataway, NJ, USA) according to the manufacturer's protocol. The filters were pre-hybridised for 5 h at 65°C in hybridisation solution (4X SSPE, 2X Denhardt's solution, 0.5% SDS, 6% Polyethyleneglycol, 40 μg/ml denatured sonicated salmon sperm DNA) before adding the denatured probe. After 18 h of hybridisation, filters were washed at 65°C, using a final wash of 0.1X SSC, 0.1% SDS. The filters were exposed to X-ray Biomax film, using standard techniques.
Densitometry
Densitometric analysis of Southern filters hybridised with EYA1 probes was performed as previously described using the Molecular Dynamics 300A Computing Densitometer (Amersham Pharmacia Biotech, Piscataway, NJ, USA).29 A single copy probe from chromosome 4 (D4S12) was hybridised to each filter as a control. Hybridisation signals of both D4S12 and the particular EYA1 probes were measured.
RNA extraction, cDNA synthesis and amplification
Total RNA was extracted from lymphoblastoid cell lines (approximately 3×106 cells) using TRIzol® LS (Life Technologies) according to the manufacturer's procedure. Samples were treated with DNase I/Amp (Life Technologies) for 15 min at room temperature and purified using the RNeasy Mini Kit (QIAGEN, Valencia, CA, USA). Approximately 3 μg of total RNA were reverse-transcribed into cDNA using random hexamers (SuperScript™ Preamplification System, Life Technologies). The cDNA templates were tested by amplification of exon 10 of the alpha subunits of IkappaB kinase (IIKα) gene, known to be ubiquitously expressed, using forward primer 5′-TCC TCG GAA ACC AGC CTC TCA ATG T-3′ in exon 9 and reverse 5′-TAA AGT GTG GGC TGA AGC AGT GCA-3′ in exon 11 under the following conditions: 35 cycles consisting of 1 min at 95°C, 1 min at 55°C, 30 s at 72°C. The last extension was lengthened to 5 min.
Amplification of the full-length EYA1 cDNA
To obtain full-length EYA1 cDNA, a 1/100 dilution of Human Foetal Kidney 5′ stretch Plus cDNA library (Clontech, Palo Alto, CA, USA) was amplified using primers ‘EYA1cDNA’ (Table 1). The PCR reaction was performed in a total volume of 50 μl, containing 20 μM of dNTPs, 10 μM of each primer and one unit of Sigma Taq polymerase. The cDNA was amplified for 35 cycles consisting of 1 min at 95°C, 1 min at 62°C, 2 min at 72°C. The last extension was lengthened to 7 min. Alignment between the known EYA1 cDNA sequence (GenBank accession no.Y10260) and the kidney cDNA sequence was performed using the program Windows 32 SeqMan 4.05© (DNASTAR Inc, Madison, WI, USA)
Inverse-PCR
Genomic DNA from an affected individual of the K6015 family was digested with PstI and electrophoresed on a 0.8% Seakem® LE agarose gel. DNA fragments between 1.6 and 2 kb were gel-purified using QIAquick gel extraction kit. Fragments were self-ligated overnight at a concentration of 3 ng/μl with one unit of T4 DNA ligase (Promega, Madison, WI, USA) and amplified using exon 1′ complementary primers (exon 1′ I-PCR) (Table 1), in order to amplify flanking sequences. The PCR reaction was performed on 1 μl of self-ligated fragments in a total volume of 30 μl, containing 10 μM of each primer, 25 μM dNTPs, 1X PCR buffer (1.5 mM MgCl2) and one unit of Taq polymerase, under the following conditions: 95°C for 4 min, 35 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 2 min, with a final extension of 7 min. The PCR product was analysed on a 1% gel and a 1.7 kb fragment was gel-purified as described above and directly sequenced.
Long-Range PCR
Long-range PCR were performed using Taq polymerase with 10X PCR Buffers 1 (17.5 mM MgCl2) and 3 (22.5 mM MgCl2 and detergents) from the Expand Long template PCR system (Roche, Indianapolis, IN, USA) following the manufacturer's recommendations.
Results
SSCP and sequence analysis
A small insertion/deletion in the K6030 family
Seventeen primer pairs were used for the amplification of EYA1 exons using genomic DNA of unaffected and affected individuals from all families. SSCP electrophoresis detected a mobility shift in exon 9 of kindred K6030 (Figure 1A). After cloning the PCR product, multiple clones were sequenced to isolate both mutated and normal alleles. A T→AG change at base 107 of exon 9 created a frameshift that causes an immediate stop codon (TAG) and truncation of the EYA1 gene product (Figure 1B). The T→AG alteration also creates a StyI restriction site, which allowed us to amplify and digest genomic DNA of all the family members. Segregation of this mutation in the K6030 family confirmed that it was linked to the BOR phenotype (data not shown).
Other than an already characterised polymorphism (C1179T) in exon 12,18 no differences between the other patients and controls could be detected by SSCP analysis. All EYA1 exons were amplified and bi-directionally sequenced from genomic DNA of the proband in all families. No additional sequence alterations were detected.
Southern analysis
A 70 bp deletion in the K6380 family
Since all PCR-based procedures failed to detect mutations in four families linked to the BOR critical region, digested genomic DNA from the four probands and two controls was hybridised with EYA1 cDNA probes to screen for deletions, duplications and complex rearrangements. Potential deletions/duplications were analysed by densitometry.
Hybridisation of a DraI Southern filter with probes EYA1.2 (exons 1 to 12) and EYA1.4 (exons 4 to 8) detected an extra fragment of about 550 bp in the proband from K6380, whereas the EYA1.1 probe (5′UTR to exon 3) did not (Figure 2A). Also, the normal exon 7 fragment (655 bp) was of lower intensity. Therefore, the shifted fragment was assumed to contain exon 7. Amplification of the patient's genomic DNA using exon 6 forward and exon 7 reverse primers, followed by a DraI digestion of the 831 bp product, confirmed the presence of a 550 bp extra fragment, which is absent in control DNA (Figure 2B). Sequencing of the 550 bp fragment revealed a 70 bp deletion starting in intron 6, 15 bp before exon 7 and extending into exon 7 (Figure 2C). Segregation of the deleted allele with the BOR phenotype was confirmed by amplification of exon 717 in seven affected and four unaffected relatives (data not shown).
Characterisation of the genomic rearrangement in kindred K6380. (A) Southern hybridisation of a DraI filter with probe EYA1.4 showing the normal 660 bp band (exon 7) and the shifted band (550 bp) (arrow) in the proband from K6380. Exon 4 (0.8 kb band) and exon 5 (1 kb band) are normal. The 290 bp band containing exon 6 did not hybridise. The bottom panel corresponds to an autosomal single copy probe D4S12. (B) Agarose gel electrophoresis of a DraI digestion of amplified genomic DNA containing exon 6 to 7 revealing an extra 550 bp band (arrow) in the K6380 proband (M=1 kb ladder). (C) Sequence alignment between normal and affected alleles, revealing a 70 bp deletion starting in intron 6, encompassing the splicing site (underlined) and part of exon 7 coding sequence.
A large deletion in the K6190 family
Hybridisation of a PstI filter with probe EYA1.1 (5′UTR to exon 3) revealed that one copy of exons 2 and 3, but not exons 1 and 1′, were probably deleted in the proband of K6190 because the fragments exhibited lower intensities (Figure 3A). Hybridisation of a DraI filter with probe EYA1.4 revealed that exons 4 to 8 also were deleted (Figure 3B). Hybridisation with probe EYA1.3 (exon 13 to 3′UTR) on a PstI filter looked normal (Figure 3C). Finally, hybridisation with probe EYA1.5 (exon 9 to 11) revealed that one copy of exon 9 was probably absent but that exon 11 was present (Figure 3D). These results were confirmed by densitometric analysis (Table 2). RT–PCR using primers in exons 1 and 13 (Table 1) allowed amplification of four products of sizes smaller than expected for the normal allele in the K6190 proband only (data not shown). After gel-purification, each amplification product was directly sequenced and revealed a deletion of exons 2 through 9. The four products corresponded to different combinations of alternative splicing of exon 1′ and 10. Amplification of a human foetal kidney cDNA library had previously identified a novel isoform of the EYA1 gene (Vervoort, unpublished data; GenBank accession no. AF467247) revealing alternative splicing of exon 10, which was consistent with the RT–PCR results. Therefore, the combined results from Southern hybridisation and RT–PCR strongly suggest a deletion of exons 2 to 9, and located the deletion breakpoint regions within intron 1′ and intron 9. Hybridisation of a HindIII filter with probe EYA1.1 allowed detection of a shifted fragment and placed the 5′end of the deletion close to exon 1′ (Figure 4A). Hybridisation with probe intron 9C allowed us to detect the same shifted fragment as detected with exon 1′, thereby identifying the 3′ end of the deletion (Figure 4B, Table 2). Using primers ‘DeletionK6190’ (Table 1) designed according to these results, it was possible to amplify a 2 kb PCR product from the three affected members of K6190, but not from a control (Figure 4C). Sequencing of the 2 kb fragment revealed it resulted from a 85 kb deletion, the 5′end breakpoint being located in intron 1′, 757 bp after the end of exon 1′ and the 3′ end breakpoint being located in intron 9, 6.9 kb downstream of exon 9 (Figure 4D). All three affected members of K6190 family had the exact same breakpoint sequence. A BLAST search (http://www.ncbi.hlm.nih.gov/blast) using sequence from the breakpoint junction fragment identified a 24 bp insertion, which was 100% homologous to a contig from the BAC clone RP11–11K9, which contains the EYA1 gene. It was not possible to determine if the 24 bp sequence was part of intron 10 or mapped upstream of the EYA1 coding sequence. This insertion was preceded by 3 bp sequence (TAG) of unknown origin.
Southern hybridisation of various filters with different EYA1 cDNA probes revealing exon deletions in the K6190 proband. (A) PstI filter hybridised with probe EYA1.1. Note the lower intensity of exon 2 (6.3 kb band) and exon 3 (4.9 kb band) fragments. Exon 1 (0.87 kb band) and 1′ (1.3 kb band) fragments appear normal. (B) DraI filter hybridised with probe EYA1.4 showing lower intensities of exon 4 (0.8 kb), exon 5 (1 kb), exon 7 (660 bp) and 8 (3 kb). (C) PstI filter hybridised with probe EYA1.3 showing that exons 13 through 15 (upper) and exon 16 through 3′UTR (lower) fragments are normal. (D) HindIII filter hybridised with probe EYA1.5 showing lower intensity of exon 9 (3.9 kb band). Exon 11 (4 kb band) is normal. An autosomal single copy probe D4S12 is displayed for each filter.
Characterisation of the genomic rearrangement in K6190 by Southern analysis. (A) Hybridisation of a HindIII filter with probe EYA1.1 showing the shifted band (2.8 kb) in the affected members only, corresponding to the 5′ end breakpoint of the deletion. (B) Hybridisation of a HindIII filter with probe Intron 9C showing lower intensity of the normal 1.8 kb and 2.2 kb bands in the affected members only and a 2.8 kb shifted band in the affected members only. The 2.8 kb shifted band corresponds to the 3′ end breakpoint of the deletion and is the same band as revealed with EYA1.1 probe in panel A. (C) Agarose gel electrophoresis showing PCR amplification of the deletion breakpoint using primers ‘deletionK6190’ on the three affected family members. The bright band in lane M (100 bp ladder) corresponds to 600 bp. (D) Schematic of the genomic structure of EYA1 showing the location of intronic probes 9B and 9C and the 5′ and 3′ end breakpoints characterised in the affected members of kindred K6190. The normal sequence flanking the breakpoint in introns 1′ and 9 are shown below, as well as the deleted allele found in all three affected individuals of the K6190 family. The underlined sequence corresponds to the 24 bp insertion homologous to BAC RP11–11K9 that contains part of the EYA1 gene. A 3 bp sequence, TAG, of unknown origin is indicated by bold characters.
A large inversion in the K6015 family
Both probes EYA1.2 (exons 1 to 12) and EYA1.1 (5′UTR to exon 3) revealed an extra 1.7 kb fragment in K6015 on a PstI filter (Figure 3A). Exons 1, 1′, 2 and 3 were hybridised individually and only exon 1′ hybridised to the extra fragment (Figure 5A). Inverse-PCR of the self-ligated PstI digested fragment, using exon 1′ complementary primers (Table 1) allowed amplification of the unknown flanking sequence. By direct sequencing of the I-PCR product, we identified a new 565 bp sequence starting 141 bp after the end of exon 1′. A BLAST search against the ‘high through genome sequence’ (htgs) database from NCBI using the new sequence identified a BAC clone, RP11–99A14 (GenBank accession no. AC022826) that mapped to chromosome 8. Using the electronic PCR option from NCBI (http://www.ncbi.nlm.nih.gov/genome/sts/epcr.cgi), nine markers were identified, including D8S1972 and D8S1172, in this BAC. These markers allowed us to map the unknown sequence about 3 cM telomeric to the EYA1 gene. Because of the inverted orientation of the new sequence compared to the BAC sequence found by I-PCR and the telomeric location of the new sequence, it was decided to test for an inversion of the entire region. By designing primers ‘K6015inversion3′1 (Table 1) from the flanking sequences of the breakpoints (intron 1′ and telomeric sequence), it was possible to amplify and sequence a 530 bp fragment, revealing a head to tail conformation, which confirmed the inversion rearrangement (Figure 5B). An 8 bp sequence ‘TGACTTAC’ from intron 1′ was found to be present at both breakpoints of the inversion in opposite directions and a 3 bp sequence (CTT) from BAC RP11–99A14 was missing (Figure 5B). The breakpoint occurred within EST sts-AA025113, 159 bp upstream of a polyadenylation site. This EST corresponds to the 3′UTR of a gene of unknown function (Unigene cluster Hs.61250) composed of four overlapping ESTs.
Characterisation of the genomic rearrangement in K6015. (A) Southern blot hybridisation using an exon 1′ probe showing an extra 1.7 kb PstI band (arrow). (B) Schematic of the normal chromosome and the affected chromosome in K6015 showing the inversion. The inverted duplicated sequence is underlined and the 3 bp deletion is in bold characters. (C) Segregation of the K6015 inversion in the EYA1 gene. Southern hybridisation of a PstI filter with RP11–99A14 probe. Note that the two extra bands (2.3 kb and 1.7 kb) are only present in the affected members of the family, which confirms segregation of the rearrangement within the family.
Southern hybridisation of a K6015 family PstI filter with a probe containing the sequence of the breakpoint region from BAC clone RP11–99A14 (Table 1) identified a second shifted fragment of 2.3 kb, along with the originally observed 1.7 kb shifted fragment, corresponding to the other end of the rearrangement that was only present in the nine affected members of the K6015 family (Figure 5C).
A complex rearrangement in the K6310 family
Hybridisation of a PstI filter with probe EYA1.1 showed exons 1, 1′ and 2 had normal intensities but exon 3 had reduced intensity in the K6310 proband (Table 2). Hybridisation of another PstI filter with probe EYA1.4 showed that exons 4 and 5 had a lower intensity but exons 6 through 8 were present in two copies (Table 2). Hybridisation of a Sau3AI filter with probe EYA1.5 showed that exon 9 was normal, but surprisingly exon 11 was less intense (Table 2). Hybridisation of a MspI filter with exon 10 and its flanking sequence showed that it also had a lower intensity (Table 2). Hybridisation of a PstI filter with probe EYA1.3 showed that exons 13 though 15 were less intense but exon 16 was normal (Table 2). And finally exon 15 and its flanking sequences were hybridised to a PstI filter and also exhibited a reduction in intensity (Table 2).
Based on these results, exons 3 through 5 and exons 10 through 15 appear to be deleted, whereas exons 1–2, 6 through 9 and exon 16 seem normal. Hybridisation with probes from introns 2, 5, 9 and 15 was performed in order to refine the four breakpoint regions (Table 3). A polymorphism was detected in intron 15, using probe intron 15B, also revealing the presence of both copies in all the affected members of the family (data not shown). Probes from intron 5 failed either to amplify or hybridize specifically, possibly due to the high content of repeat sequences. Since none of the other probes detected any shift, long-range PCR was performed to try to amplify the breakpoints: a possible large deletion with an insertion of exons 6 through 9 outside the coding region; a potential existence of two independent deletions between introns 2 and 5 and between introns 9 and 15; a possible inversion associated with a deletion. Unfortunately, no PCR product was generated in any of these reactions. Thus, although there was good evidence that the EYA1 gene was disrupted in family K6310, it was not possible to fully characterise this complex rearrangement.
Discussion
Several mutation reports have confirmed that alterations in the human homologue of the Drosophila eyes absent gene, EYA1, cause BO/BOR syndrome.17,18,19,20,21,2223 However, in some of these reports only 20% of the patients studied had confirmed mutations in the EYA1 gene. To explain a low mutation detection rate, some investigators have hypothesised that mutations in another gene tightly linked to EYA1 also cause BOR syndrome.26 However, using Southern analysis it was possible to detect mutations in our panel of four families linked to chromosome 8 in which SSCP analysis and direct sequencing had failed to detect a point mutation. This underscores the major drawback of many PCR-based mutation detection procedures in the study of an autosomal dominant disorder which is the inevitable amplification of the normal allele, which can mask deleted or disrupted exons.30,31
A deletion of 85 kb was identified in family K6190, which has a typical BOR phenotype. This deletion appears to be the largest intragenic deletion thus far reported for BO/BOR syndrome. Unlike most of the reported deletions in which the translated protein would be truncated for the eyaHR domain,17,18,19,20,2122 this deletion eliminates half of the exons in the variable region of EYA1 leaving most of the eyaHR domain intact. The truncated EYA1 mRNA is apparently quite stable since it was possible to amplify four truncated cDNA isoforms from this patient's lymphoblasts. This finding shows that mutations outside the highly conserved domain eyaHR can give rise to the BOR phenotype.
The large paracentric inversion characterised in family K6015 is the first reported inversion that disrupts the entire EYA1 gene. One of the breakpoints is located at the 5′ end of intron 1′ and the other breakpoint is located outside of the EYA1 gene. This mutation indicates that if Southern analysis is used to detect rearrangements, multiple cDNA or genomic probes should be utilised since in this case most of the gene (16 exons out of 17) appeared normal by Southern analysis. Indeed, the presumed rearrangement in another family, K6310, could not be completely characterised, indicating that even Southern analysis is not capable of resolving all complex rearrangements. Although no shifted fragment could be detected, dosage analysis revealed that exons 3 to 5 and exons 10 to 15 were deleted, while exons 6 to 9 appeared normal.
Some initial reports on BOR syndrome describing one patient with a dir ins(8)(q24.11;q13.3;q21.13)15,32 and another patient with 8q12.2-q21 deletion11 indicated that complex genomic rearrangements can cause a BOR phenotype. In fact, in the first two mutation reports, three large deletions of 7, 5.6 and 20–37 kb are described.17,18 Including the rearrangements reported here, eight complex rearrangements, corresponding to about 21% (8/38) of published BOR mutations, have been identified. This frequency of complex rearrangements may indicate that the EYA1 region is unstable.
The only mutation identified by SSCP and sequencing was a small insertion/deletion in exon 9 (T917AG) in K6030. It is interesting to note that K6030 is the second case report of a BO family with a frameshift mutation in exon 9.23 Several clinical reports have previously suggested that the BO syndrome was allelic to the BOR syndrome, since silent renal anomalies,33 underdiagnosed kidney problems34 and severe renal manifestations are only expected in 6% of BOR patients.4 K6030 is now the sixth BO family associated with EYA1 mutations.22,23
In conclusion, using PCR-based methods, such as SSCP and sequencing, only one point mutation in EYA1 was detected in 5 BOR families linked to chromosome 8q13.3. However, using Southern blot analysis, complex gene alterations including a large inversion and two deletions were found in the other four families. A high proportion (21%) of complex genomic rearrangements involving the EYA1 gene have been reported. However, these rearrangements can not be detected by commonly used mutation screening procedures and these data suggest that more complex rearrangements remain to be detected in BOR patients with missing mutation in EYA1. Although most mutations result in truncated proteins, the etiology is quite different for each patient, from a single base change to complex genomic rearrangements that make it impossible to use a single approach for mutation screening of EYA1. The use of Southern analysis or quantitative PCR in combination with SSCP should cover a wide range of mutations found in the EYA1 gene.
References
Melnick M, Bixler D, Silk K, Yune H, Nance WE . Autosomal dominant branchiootorenal dysplasia Birth Defects Orig Art Ser 1975 XI (5): 121–128
Melnick M, Bixler D, Nance WE, Silk K, Yune H . Familial Branchio-Oto-Renal dysplasia: a new addition to the branchial arch syndromes Clin Genet 1976 9: 25–34
Fraser FC, Ling D, Clogg D, Nogrady B . Genetic aspects of the BOR syndrome branchial fistulas, ear pits, hearing loss, and renal anomalies. Am J Med Genet 1978 2: 241–252
Fraser FC, Sproule JR, Halal F . Frequency of the Branchio-Oto-Renal (BOR) syndrome in children with profound hearing loss Am J Med Genet 1980 7: 341–349
Cremers CWRJ, Fikkers-van Noord M . The earpits-deafness syndrome: clinical and genetic aspects Int J Pediatr Otorhinolaryngol 1980 2: 309–322
Chen A, Francis M, Ni L et al.. Phenotypic manifestations of Branchio-Oto-Renal syndrome Am J Med Genet 1995 58: 365–370
Kumar S, Kimberling WJ, Kenyon JB, Smith RJ, Marres HA, Cremers CW . Autosomal dominant Branchio-Oto-Renal syndrome localization of a disease gene to chromosome 8q by linkage in a Dutch family Hum Mol Genet 1992 1: 491–495
Smith RJ, Coppage KB, Ankerstjerne JK et al. Localization of the gene for Branchio-Oto-Renal syndrome to chromosome 8q Genomics 1992 14: 841–844
Wang Y, Treat K, Schroer RJ, O'Brien JE, Stevenson RE, Schwartz CE . Genome, Localization of Branchio-Oto-Renal (BOR) syndrome to a 3 Mb region of chromosome 8q Am J Med Genet 1994 51: 169–175
Kumar S, Kimberling WJ, Connolly CJ, Tinley S, Marres HA, Cremers CW . Refining the region of Branchio-Oto-Renal syndrome and defining the flanking markers on chromosome 8q by genetic mapping Am J Hum Genet 1994 55: 1188–1194
Vincent C, Kalatzis V, Compain S et al. A proposed new contiguous gene syndrome on 8q consists of Branchio-Oto-Renal (BOR) syndrome, Duane syndrome, a dominant form of hydrocephalus and trapeze aplasia; implications for the mapping of the BOR gene Hum Mol Genet 1994 3: 1859–1866
Ni L, Wagner MJ, Kimberling WJ et al. Genome, Refined localization of the branchiootorenal syndrome gene by linkage and haplotype analysis Am J Med Genet 1994 51: 176–184
Kumar S, Kimberling WJ, Lanyi A et al. Narrowing the genetic interval and yeast artificial chromosome map in the Branchio-Oto-Renal region on chromosome 8q Genomics 1996 31: 71–79
Kalatzis V, Abdelhak S, Compain S, Vincent C, Petit C . Characterization of a translocation-associated deletion defines the candidate region for the gene responsible for Branchio-Oto-Renal syndrome Genomics 1996 34: 422–425
Gu JZ, Wagner MJ, Haan EA, Wells DE . Detection of a megabase deletion in a patient with Branchio-Oto-Renal syndrome (BOR) and tricho-rhino-phalangeal syndrome (TRPS): implications for mapping and cloning of the BOR gene Genomics 1996 31: 201–206
Kumar S, Deffenbacher K, Marres HA et al. Genomewide search and genetic localization of a second gene associated with autosomal dominant branchio-oto-renal syndrome: clinical and genetic implications Am J Hum Genet 2000 66: 1715–1720
Abdelhak S, Kalatzis V, Heilig R et al. Protein, Nucleotide, A human homologue of the Drosophila eyes absent gene underlies Branchio-Oto-Renal (BOR) syndrome and identifies a novel gene family Nature Genet 1997 15: 157–164
Abdelhak S, Kalatzis V, Heilig R et al. Protein, Nucleotide, Clustering of mutations responsible for Branchio-Oto-Renal (BOR) syndrome in the eyes absent homologous region (eyaHR) of EYA1 Hum Mol Genet 1997 6: 2247–2255
Kumar S, Kimberling WJ, Weston MD et al. Identification of three novel mutations in human EYA1 protein associated with Branchio-Oto-Renal syndrome Hum Mutat 1998 11: 443–449
Kumar S, Deffenbacher K, Cremers CW, Van Camp G, Kimberling WJ . Branchio-Oto-Renal syndrome: identification of novel mutations, molecular characterization, mutation distribution, and prospects for genetic testing Genet Test 1998 1: 243–251
Usami S, Abe S, Shinkawa H, Deffenbacher K, Kumar S, Kimberling WJ . EYA1 nonsense mutation in a Japanese Branchio-Oto-Renal syndrome family J Hum Genet 1999 44: 261–265
Rickard S, Boxer M, Trompeter R, Bitner-Glindzicz M . Importance of clinical evaluation and molecular testing in the Branchio-Oto-Renal (BOR) syndrome and overlapping phenotypes J Med Genet 2000 37: 623–627
Vincent C, Kalatzis V, Abdelhak S et al. BOR and BO syndromes are allelic defects of EYA1 Eur J Hum Genet 1997 5: 242–246
Bonini NM, Bui QT, Gray-Board GL, Warrick JM . The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates Development 1997 124: 4819–4826
Azuma N, Hirakiyama A, Inoue T, Asaka A, Yamada M . Mutations of a human homologue of the Drosophila eyes absent gene (EYA1) detected in patients with congenital cataracts and ocular anterior segment anomalies Hum Mol Genet 2000 9: 363–366
Lin X, Wells DE, Kimberling WJ, Kumar S . Human NDUFB9 gene: genomic organization and a possible candidate gene associated with deafness disorder mapped to chromosome 8q13 Hum Hered 1999 49: 75–80
Schwartz CE, Ulmer J, Brown A, Pancoast I, Goodman HO, Stevenson RE . Allan-Herndon syndrome. II. Linkage to DNA markers in Xq21 Am J Hum Genet 1990 47: 454–458
Bolton BJ, Spurr NK . B-Lymphocytes; in Freshney R Ian, Freshney Mary G (eds) Culture of Immortalized Cells New York: Weiley-Liss 1996 pp 283–298
Tharapel AT, Anderson KP, Simpson JL et al. Deletion (X)(q26.1→q28) in a proband and her mother: molecular characterization and phenotypic-karyotypic deductions Am J Hum Genet 1993 52: 463–471
Papadopoulos N, Leach FS, Kinzler KW, Vogelstein B . Monoallelic mutation analysis (MAMA) for identifying germline mutations Nature Genet 1995 11: 99–102
Yan H, Papadopoulos N, Marra G et al. Conversion of diploidy to haploidy Nature 2000 403: 723–724
Haan EA, Hull YJ, White S, Cockinton R, Charlton P, Callen DF . Tricho-rhino-phalangeal and branchio-oto syndromes in a family with an inherited rearrangement of chromosome 8q Am J Med Genet 1989 32: 490–494
Heimler A, Lieber E . Branchio-Oto-Renal syndrome: reduced penetrance and variable expressivity in four generations of a large kindred Am J Med Genet 1986 25: 15–27
Chitayat D, Hodgkinson KA, Chen MF, Haber GD, Nakishima S, Sando I . Branchio-Oto-Renal syndrome: further delineation of an underdiagnosed syndrome Am J Med Genet 1992 43: 970–975
Acknowledgements
We wish to thank the patients for their cooperation. S Daniels of the JC Self Research Institute core facility did the sequencing and T Moss maintained the patients' cell lines. This study was supported, in part, by a grant from the South Carolina Department of Disabilities and Special Needs.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vervoort, V., Smith, R., O'Brien, J. et al. Genomic rearrangements of EYA1 account for a large fraction of families with BOR syndrome. Eur J Hum Genet 10, 757–766 (2002). https://doi.org/10.1038/sj.ejhg.5200877
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200877
Keywords
This article is cited by
-
From clinical to molecular diagnosis: relevance of diagnostic strategy in two cases of branchio-oto-renal syndrome – case report
Italian Journal of Pediatrics (2022)
-
Recurrent 8q13.2-13.3 microdeletions associated with Branchio-oto-renal syndrome are mediated by human endogenous retroviral (HERV) sequence blocks
BMC Medical Genetics (2014)
-
Branchio-oto-renal syndrome caused by partial EYA1 deletion due to LINE-1 insertion
Pediatric Nephrology (2010)
-
Branchio–oto–renal syndrome: detection of EYA1 and SIX1 mutations in five out of six Danish families by combining linkage, MLPA and sequencing analyses
European Journal of Human Genetics (2007)
-
A gene locus for progressive familial heart block type II (PFHBII) maps to chromosome 1q32.2-q32.3
Human Genetics (2005)