Abstract
Hypospadias is a congenital anomaly of the genitalia characterized by abnormalities of the urethra and foreskin, with the urethral meatus located in an abnormal position anywhere from the distal ventral penile shaft to the perineum. Because the incidence of hypospadias is approximately 1/200–1/300 live male births, it is one of the most common congenital malformations, but its etiology is largely uncharacterized. Genomic analysis of hypospadic tissue indicated a potential role for activating transcription factor 3 (ATF3) in the development of this anomaly. ATF3 may be involved in homeostasis, wound healing, cell adhesion, or apoptosis, and normally it is expressed at a steady-state in quiescent cells. Additionally, it has been shown to be an estrogen-responsive gene, and the etiology of hypospadias may be related to in utero exposure to estrogenic or anti-androgenic compounds. We examined the expression of ATF3 in tissues from 28 children with hypospadias compared with 20 normal penile skin tissue samples from elective circumcision. Eighty-six percent of the hypospadias samples were immunohistochemically positive, compared with 13% of normal tissue samples. Seventy-five percent of hypospadias samples were positive from in situ hybridization, compared with 1% of circumcision samples. Our results indicate that ATF3 is up-regulated in the penile skin tissues of boys with hypospadias, suggesting a role for this transcription factor in the development of this abnormality. Because the etiology of hypospadias may include exposure to estrogenic compounds, the responsiveness of ATF3 to estrogen is also discussed.
Similar content being viewed by others
Main
Hypospadias is a congenital anomaly of the genitalia characterized by abnormalities of the urethra and foreskin (1). In mild hypospadias the urethral meatus is located in an abnormal position along the distal ventral penile shaft, coronal margin, or proximal glans. In moderate hypospadias the abnormal urethral opening exists on the middle of ventral aspect of the penis. Severe hypospadias is characterized by a proximal urethral opening that can occur at the penile scrotal junction or within the scrotal folds or perineum, and the more severe forms of hypospadias are associated with penile curvature. The incidence of hypospadias is approximately 1/200∼1/300 live male births, making hypospadias one of the most common congenital malformations in children. Some research has documented that the incidence of this anomaly has been increasing in the United States during the last three decades (2). The molecular events required in the genitourinary tract for normal development of the male external genitalia are just beginning to be elucidated (3–6), and the etiology of hypospadias remains unknown, although endocrine disruptors have been proposed as a possible explanation for the increasing incidence that has been reported in industrialized countries (4–7). Many studies also suggest a genetic component in the transmission of this birth defect and it seems to be multifactorial (8,9) 5,5A.
Activating transcription factor 3 (ATF3) is a member of the ATF/CREB (cAMP responsive element binding) family of transcription factors (10). ATF3 contains the basic region and leucine zipper (bZIP) motif characteristic of the bZIP superfamily of transcription factors, which includes members of the CCAAT/enhancer-binding protein family, the Jun/Fos family and ATF/CREB family (10–12). Expression of ATF3 protein is normally at a steady state in quiescent cells, but can be rapidly induced to a high level in response to multiple extracellular signals, including growth factors, cytokines, and genotoxic stress agents (10,13–17).
ATF3 may be involved in homeostasis, wound healing, cell adhesion, cancer cell invasion, apoptosis and signaling pathways (10,11,18–20). Overexpression of ATF3 protein suppresses cell growth and slows the transition of cells from G1 to S phase. This evidence suggests that the ATF3 protein may play a negative role in cell cycle progression (20–23).
Microarray results from our lab (Z. Wang et al., manuscript in preparation) indicate that ATF3 is one of the genes that is up-regulated in hypospadic tissue. Exploration of the literature revealed a potential response of ATF3 to estrogen (9,24), and estrogenic or anti-androgenic activity is implicated in the development of hypospadias (4,25). To investigate further the role of ATF3 in this anomaly, we compared the gene's mRNA/protein expression in normal versus hypospadic human tissue using molecular localization techniques.
MATERIALS AND METHODS
Young males with hypospadias scheduled to undergo surgical repair were prospectively entered into this study from January, 2004 to June, 2004. We received written, informed consent from parents for these procedures, which were approved by our institutional Committee on Human Research.
Patients with an undescended testis, intersex condition, or known endocrine abnormalities were excluded from the study. The position of the urethral meatus (Table 1), associated anomalies, and family history of hypospadias were assessed by history and physical exam. No patients received preoperative testosterone treatment.
Excess prepucial tissue was obtained at the time of elective surgery for hypospadias; no periurethral tissue was taken. Controls consisted of patients undergoing elective circumcision, and controls and hypospadias samples were strictly age-matched. Tissue specimens were prepared for paraffin-embedded or frozen sectioning.
Antibodies and immunohistochemical techniques.
Skin specimens were evaluated for expression of ATF3 protein by immunohistochemical staining using the standard protocol accompanying the ABC kit (Vector Laboratories, Inc., CA), with overnight primary-antibody incubation with rabbit-anti-ATF3 (Santa Cruz Biotechnology, Inc., CA) at a dilution of 1:400. Control sections were incubated without primary antibody. Digital images were acquired with a Leica camera and ACT-1 computer software.
Preparation of ATF3 probe and in situ hybridization.
Expression of ATF3 mRNA was detected by in situ hybridization with digoxygenin-labeled ATF3 antisense probe. Total RNA was isolated from a patient (6 mo) with severe hypospadias. Reverse-transcriptase (RT) polymerase chain reaction (PCR) was performed with 2.5 μg of RNA sample in a reaction volume of 20 μL using a DNA Engine® Thermal Cycler. Fragments of the PCR product were harvested using the GeneClean method (Geneclean Kit, Qbiogene, Inc., Carlsbad, CA).
Construction of pCR 2.1-ATF3 and plasmid assay.
Ligation fragment of ATF3 with pCR 2.1 vector was processed using the instruction manual from TA cloning kit (Invitrogen, TA Cloning kit with pCR 2.1 vector). The plasmid of pCR 2.1-ATF3 was transferred into DH5αINVaF' competent cells (Invitrogen, one shot DH5αINVaF' chemically competent E. coli), followed by spreading 50 μL of the competent cells on a labeled LB agar plate containing 100 μg/mL ampicillin and X-Gal (including IPTG) and incubated overnight at 37°C. Simultaneously, the ligation reaction without X-gal was performed as a control. The white colonies were those cells successfully transfected with pCR 2.1-ATF3. The blue colonies were negative pCR 2.1-ATF3. Both white and blue clones were selected and transferred to an ampicillin LB agar plate. To increase the amount of plasmid, both colonies were cultured separately at 225 rpm for 14 h in a shaking incubator. DNA purification of plasmid was performed using Qiagen plasmid mini kit 25 (Qiagen). Plasmid DNA sequence analysis was completed by restriction enzyme digestion. Based on the nucleotide sequence of pCR2.1-ATF3, a pair of restriction endonuclease analysis for the plasmid DNA was designed by restriction enzyme EcoR-1 (BioLabs Inc.) and Xbal-1 (BioLabs Inc.) respectively. A 254 bp fragment was harvested after EcoR-1 digestion with Multi-coreTM buffer, and 350 bp fragment by Xbal-1 with Nebuffer 2.
Construction of pCR 2.1-ATF3 and plasmid assay was done as previously described (26). Based on the nucleotide sequence of pCR2.1-ATF3, a pair of restriction endonuclease enzymes (EcoR1 and Xbal-1) (BioLabs Inc.) was used to harvest a 254 bp fragment (EcoR1) and a 350 bp fragment (Xbal1).
ATF3 probe labeling.
Probe was labeled with DIG-11-UPT (DIG RNA Labeling mix, Roche) in a T7/Sp6 polymerase (Promega) transcription reaction, and appropriate signal was checked by dot blot. ATF3 DNA to be labeled was cloned into pCR 2.1, the polylinker site of an appropriate transcription vector, which contains adjacent to the polylinker a promoter for T7 RNA polymerase and Sp6 RNA polymerase. Plasmid DNA was linearized by the restriction enzymes SpeI and NotI for the synthesis of “run off” transcripts. Restriction enzymes creating 5′-overhangs were used. The linearized template DNA was purified by the gene clean method with glass beads, avoiding RNase contamination. Dig RNA Labeling Mix (10× conc., Roche) was used for ATF3 probe labeling. DIG-labeled, single-stranded RNA probes of defined length were generated by in vitro transcription. DIG-11-UPT was incorporated by Sp6 or T7 RNA polymerases at approximate every 20–25th nucleotide of the transcript under the conditions as described in the manual. The reaction system included 5× transcription buffer (T7) (Promega), 5× transcription buffer (Sp6), (Promega), 0.1 DTT (Invitrogen), Sp6 RNA polymerase (Promega), T7 RNA polymerase (Promega), DNase 1 RNasin free (Roche), RNasin inhibitor (Promega). Probe signal was checked by dot probe on a nylon membrane using UV cross-linking. Then anti-digoxygenin immunohistochemistry was performed with blocking the membrane in 500–800 μL of MABT/2% BM blocking reagent/10% sheep serum. The membrane was washed and the probe detected with BM purple.
In situ hybridization.
In situ hybridization followed standard laboratory protocols. All steps were conducted under RNase-free conditions. Frozen tissues were sectioned at 4 μm on a Zeiss HM500 cryostat. All sections were hybridized with ATF3 RNA probe at 65°C overnight in a humidified chamber. Anti-digoxygenin (1:200) immunohistochemistry was performed by using anti-dig antibody (anti-digoxygenin-AP Fab fragments, Roche Diagnostics Corporation, Indianapolis, IN, USA). BM purple was used for staining. The specificity of the ATF3 hybridization signal was verified by comparison with the sections processed with sense probe under identical conditions
Statistical analysis.
Fisher's exact two-tailed tests were used to compare the frequency data of the two groups. Statistical significance was assigned at p < 0.05.
RESULTS
Expression of ATF3 protein in prepuce.
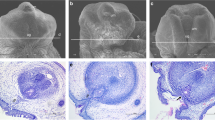
ATF3 protein localized in the nuclei of stromal cells and the vascular endothelium in the s.c. tissue of prepuce. The staining intensities were stronger in patients with moderate and severe hypospadias compared with mild hypospadias (Fig. 1, Table 2). Of the 43 specimens examined by immunohistochemistry, 24 (85.7%) of the 28 patients with hypospadias showed expression of ATF3 protein, and only 2 (13.3%) of the 15 patients undergoing circumcision were positive (p < 0.0001) (Table 2).
ATF3 immunoreactivity in hypospadic and control tissues. Figs. 1A and 1B show moderate and severe degrees of hypospadias, respectively. Fig. 1C shows circumcision control. Fig. 1D shows negative control (no primary antibody exposure). Tissue sections in Figs. 1A1, 1B1, 1C1,and 1D1 are hematoxylin/eosin stained. ATF3 is clearly expressed in the nuclei of many cells in the s.c. tissue of hypospadic samples (Figs. 1A2, 3, 4, 1B2, 3, 4, arrows). Note that there is no ATF3 immunoreactivity in circumcision control tissue (Figs. 1C2, 3, 4) or in the negative control (Figs. 1D2, 3, 4). Magnifications from top row to bottom are 100, 100, 200, and 400×, respectively. Scale bars for each row are found in far-left panel.
Expression of ATF3 mRNA in prepuce.
ATF3 mRNA signal localized in the cytoplasm of the stromal cells in the s.c. tissue of prepuce (Fig. 2). Of the 28 specimens examined by in situ hybridization, 15 (75%) of the 20 patients with hypospadias and only 1 (6%) of the eight circumcision patients exhibited up-regulation of ATF3 mRNA (p = 0.004) (Table 3).
Expression of ATF3 mRNA in tissues of hypospadic and control tissues. Fig. 2E shows hypospadias. Fig. 2F shows circumcision control. Fig. 2G shows negative control (exposure to sense probe). Tissue sections in Figs. 2E1, 2F1,and 2G1 are hematoxylin/eosin stained. Figs. 2E2 and 3 show up-regulation of ATF3 expression in hypospadias (arrows). Figs. 2F2 and 3, and 2G2 and 3 show absence of ATF3 probe binding in circumcision and negative controls. Magnifications from top row to bottom are 100, 100, 200, and 400×, respectively. Scale bars for each row are found in far-left panel.
Consistency of expression of ATF3 protein and mRNA in prepuce.
Of the 20 specimens examined both by immunohistochemistry and in situ hybridization, 8 (40%) were positive and 6 (30%) were negative simultaneously (Table 4) (p = 0.07).
DISCUSSION
Many different laboratories indicate that ATF3 is a stress-inducible gene; its mRNA level greatly increases upon exposure of cells to stress signals (14,16). Over the past decade, the physiologic significance of ATF3 dimers has not been made clear. Thus far, the main clues for the physiologic function of ATF3 come from the expression pattern in both animal models and cultured cells. The general view is that dimer formation can alter DNA binding specificity and transcriptional activities, thus expanding the ability of these bZip proteins to regulate gene expression. The ATF3 homodimer is a transcriptional repressor (20,22,23,27); however, the heterodimeric complex of ATF3 with c-Jun (or Jun D) has been demonstrated to function as a transcriptional activator (28). Currently, our understanding in these areas is limited. It is almost certain that each ATF/CREB protein regulates a variety of target genes via complex mechanisms. Many publications have implicated ATF3 in several processes including stress response, hepatitis B viral-mediated processes, and tumorigenicity (10,16,17,20).
To our knowledge, this study is the first to demonstrate a relationship between ATF3 and hypospadias. We have shown that the ATF3 gene is up-regulated in the skin of patients with hypospadias compared with normal prepuce. Research employing a gain-of-function approach found that expression of ATF3 leads to adverse outcomes in cells exposed to stress (29). This finding is consistent with the observation that transgenic mice expressing ATF3 in selective tissues have a malfunction in target tissues (29). Although the functional significance of ATF3 up-regulation is unknown, our results suggest that ATF3 may play a role in hypospadias, not as a protective response for the cells to cope with stress, but as a part of the cellular response that leads to hypospadias formation.
Studies using the tetracycline-inducible system (Tet-off) found that over-expression of ATF3 protein suppresses cell growth (30). The results of these studies indicate that regulation of ATF3 induction after DNA damage utilizes both the p53-dependent and -independent pathways, and may also involve mitogen-activated protein kinase signaling pathways. Over-expression of ATF3 protein can slow progression of cells from G1 to S phase, indicating that ATF3 protein might play a negative role in the control of cell cycle progression (30). We suggest that over-expression of ATF3 protein in the s.c. tissue of hypospadias can inhibit formation of male normal genital tubercle and/or urethral seam. In the present study, both the protein and mRNA of ATF3 are expressed in the s.c. tissue of patients with hypospadias. The up-regulation of ATF3 in the stroma is consistent with known epithelial-stromal interactions that are critical in the development of genitalia (31).
ATF3 has been shown to be influenced by estrogen (9,24). We have previously shown that in mice, the synthetic estrogen 17-α ethinyl estradiol can cause hypospadias in the male offspring of pregnant females exposed to it (4,8,32,33). It is possible that ATF3 may regulate estrogen-responsive genes in the development process of hypospadias. Our quantitative RT-PCR work with the mouse model indicates that ATF3 is significantly up-regulated at a critical stage in urethral closure in the genital tubercles of mice that have been exposed to estrogen in utero and that it is more highly expressed in females (B. Liu et al., in preparation). Quantitative RT-PCR work with human tissue shows that ATF3 mRNA is up-regulated in hypospadic tissue compared with normal tissue (B. Liu et al., in preparation).
Further investigation of ATF3 in isolated hypospadias, including a more thorough endocrine evaluation of affected patients, would be helpful for proving a causal relationship between ATF3 gene and hypospadias. Other investigations of in vitro expression, in vivo regulation, and tissue distribution of ATF3 in the fetal genital tubercle would be helpful to clarify how ATF3 contributes to the development of hypospadias.
Abbreviations
- ATF3:
-
activating transcription factor 3
- bZIP:
-
basic region and leucine zipper
- CREB:
-
cAMP responsive element binding
- IHC:
-
immunohistochemistry
- ISH:
-
in situ hybridization
References
Baskin LS 2004 Hypospadias. Adv Exp Med Biol 545: 3–22
Paulozzi LJ, Lary JM 1999 Laterality patterns in infants with external birth defects. Teratology 60: 265–271
Baskin LS, Erol A, Jegatheesan P, Li Y, Liu W, Cunha GR 2001 Urethral seam formation and hypospadias. Cell Tissue Res 305: 379–387
Baskin LS, Himes K, Colborn T 2001 Hypospadias and endocrine disruption: is there a connection?. Environ Health Perspect 109: 1175–1183
Baskin LS, Liu W, Bastacky J, Yucel S 2004 Anatomical studies of the mouse genital tubercle. Adv Exp Med Biol 545: 103–121
Steinhardt GF 2004 Endocrine disruption and hypospadias. Adv Exp Med Biol 545: 203–215
Cohn M 2004 Developmental genetics of hypospadias. In: Baskin LS (eds) Advances in experimental medicine and biology Hypospadias and genital development. Kluwer Academic/Plenum Publishers, New York, pp 149–158
Baskin LS 2000 Hypospadias and urethral development. J Urol 163: 951–956
Nordenskjold A, Friedman E, Tapper-Persson M, Soderhall C, Leviav A, Svensson J, Anvret M 1999 Screening for mutations in candidate genes for hypospadias. Urol Res 27: 49–55
Inoue A, Yoshida N, Omoto Y, Oguchi S, Yamori T, Kiyama R, Hayashi S 2002 Development of cDNA microarray for expression profiling of estrogen-responsive genes. J Mol Endocrinol 29: 175–192
Hai T, Hartman MG 2001 The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene 273: 1–11
Cai Y, Zhang C, Nawa T, Aso T, Tanaka M, Oshiro S, Ichijo H, Kitajima S 2000 Homocysteine-responsive ATF3 gene expression in human vascular endothelial cells: activation of c-Jun NH(2)-terminal kinase and promoter response element. Blood 96: 2140–2148
Allen-Jennings AE, Hartman MG, Kociba GJ, Hai T 2002 The roles of ATF3 in liver dysfunction and the regulation of phosphoenolpyruvate carboxykinase gene expression. J Biol Chem 277: 20020–20025
Hashimoto Y, Zhang C, Kawauchi J, Imoto I, Adachi MT, Inazawa J, Amagasa T, Hai T, Kitajima S 2002 An alternatively spliced isoform of transcriptional repressor ATF3 and its induction by stress stimuli. Nucleic Acids Res 30: 2398–2406
Dussor GO, Price TJ, Flores CM 2003 Activating transcription factor 3 mRNA is upregulated in primary cultures of trigeminal ganglion neurons. Brain Res Mol Brain Res 118: 156–159
Kool J, Hamdi M, Cornelissen-Steijger P, van der Eb AJ, Terleth C, van Dam H 2003 Induction of ATF3 by ionizing radiation is mediated via a signaling pathway that includes ATM Nibrin1 stress-induced MAPkinases and ATF-2. Oncogene 22: 4235–4242
Pan Y, Chen H, Siu F, Kilberg MS 2003 Amino acid deprivation and endoplasmic reticulum stress induce expression of multiple activating transcription factor-3 mRNA species that when overexpressed in HepG2 cells, modulate transcription by the human asparagine synthetase promoter. J Biol Chem 278: 38402–38412
Hunt D, Hossain-Ibrahim K, Mason MR, Coffin RS, Lieberman AR, Winterbottom J, Anderson PN 2004 ATF3 upregulation in glia during Wallerian degeneration: differential expression in peripheral nerves and CNS white matter. BMC Neurosci 5: 9–20
Wolfgang CD, Liang G, Okamoto Y, Allen AE, Hai T 2000 Transcriptional autorepression of the stress-inducible gene ATF3. J Biol Chem 275: 16865–16870
Allen-Jennings AE, Hartman MG, Kociba GJ, Hai T 2001 The roles of ATF3 in glucose homeostasis A transgenic mouse model with liver dysfunction and defects in endocrine pancreas. J Biol Chem 276: 29507–29514
Kawauchi J, Zhang C, Nobori K, Hashimoto Y, Adachi MT, Noda A, Sunamori M, Kitajima S 2002 Transcriptional repressor activating transcription factor 3 protects human umbilical vein endothelial cells from tumor necrosis factor-alpha-induced apoptosis through down-regulation of p53 transcription. J Biol Chem 277: 39025–39034
Ishiguro T, Nagawa H, Naito M, Tsuruo T 2000 Inhibitory effect of ATF3 antisense oligonucleotide on ectopic growth of HT29 human colon cancer cells. Jpn J Cancer Res 91: 833–836
Nobori K, Ito H, Tamamori-Adachi M, Adachi S, Ono Y, Kawauchi J, Kitajima S, Marumo F, Isobe M 2002 ATF3 inhibits doxorubicin-induced apoptosis in cardiac myocytes: a novel cardioprotective role of ATF3. J Mol Cell Cardiol 34: 1387–1397
Zhang C, Gao C, Kawauchi J, Hashimoto Y, Tsuchida N, Kitajima S 2002 Transcriptional activation of the human stress-inducible transcriptional repressor ATF3 gene promoter by p53. Biochem Biophys Res Commun 297: 1302–1310
Pedram A, Razandi M, Aitkenhead M, Hughes CC, Levin ER 2002 Integration of the non-genomic and genomic actions of estrogen. Membrane-initiated signaling by steroid to transcription and cell biology. J Biol Chem 277: 50768–50775
Reference Not Provided.
Koga A, Hori H, Sakaizumi M 2002 Gene transfer and cloning of flanking chromosomal regions using the medaka fish Tol2 transposable element. Mar Biotechnol NY 4: 6–11
Ishiguro T, Nagawa H 2000 Expression of the ATF3 gene on cell lines and surgically excised specimens. Oncol Res 12: 181–183
Tsujino H, Kondo E, Fukuoka T, Dai Y, Tokunaga A, Miki K, Yonenobu K, Ochi T, Noguchi K 2000 Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: a novel neuronal marker of nerve injury. Mol Cell Neurosci 15: 170–182
Okamoto Y, Chaves A, Chen J, Kelley R, Jones K, Weed HG, Gardner KL, Gangi L, Yamaguchi M, Klomkleaw W, Nakayama T, Hamlin RL, Carnes C, Altschuld R, Bauer J, Hai T 2001 Transgenic mice with cardiac-specific expression of activating transcription factor 3 a stress-inducible gene have conduction abnormalities and contractile dysfunction. Am J Pathol 159: 639–650
Fan F, Jin S, Amundson SA, Tong T, Fan W, Zhao H, Zhu X, Mazzacurati L, Li X, Petrik KL, Fornace AJ Jr, Rajasekaran B, Zhan Q 2002 ATF3 induction following DNA damage is regulated by distinct signaling pathways and over-expression of ATF3 protein suppresses cells growth. Oncogene 21: 7488–7496
Kurzrock EA, Baskin LS, Li Y, Cunha GR 1999 Epithelial-mesenchymal interactions in development of the mouse fetal genital tubercle. Cells Tissues Organs 164: 125–130
Yucel S, Cavalcanti AG, Desouza A, Wang Z, Baskin LS 2003 The effect of oestrogen and testosterone on the urethral seam of the developing male mouse genital tubercle. BJU Int 92: 1016–1021
Kim KS, Torres CR Jr, Yucel S, Raimondo K, Cunha GR, Baskin LS 2004 Induction of hypospadias in a murine model by maternal exposure to synthetic estrogens. Environ Res 94: 267–275
Acknowledgements
The authors thank Dr. Ching-Shwun Lin for many helpful discussions. Konijeti Ramdev helped with sample collections.
Author information
Authors and Affiliations
Corresponding author
Additional information
This study was supported by NIH grant DK 058105-01 and the American Foundation of Urologic Disease.
Rights and permissions
About this article
Cite this article
Liu, B., Wang, Z., Lin, G. et al. Activating Transcription Factor 3 Is Up-Regulated in Patients with Hypospadias. Pediatr Res 58, 1280–1283 (2005). https://doi.org/10.1203/01.pdr.0000187796.28007.2d
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1203/01.pdr.0000187796.28007.2d
This article is cited by
-
Current understanding of hypospadias: relevance of animal models
Nature Reviews Urology (2015)
-
Increased gene copy number of VAMP7 disrupts human male urogenital development through altered estrogen action
Nature Medicine (2014)
-
A pilot study of the association between genetic polymorphisms involved in estrogen signaling and infant male genital phenotypes
Asian Journal of Andrology (2012)
-
Candidate genes and their response to environmental agents in the etiology of hypospadias
Nature Clinical Practice Urology (2007)