Abstract
Purpose: Prenatal screening for Down syndrome has improved, but the number of resulting invasive diagnostic procedures remains problematic. Measurement of circulating cell-free DNA in maternal plasma might offer improvement.
Methods: A blinded, nested case-control study was designed within a cohort of 4664 pregnancies at high risk for Down syndrome. Fetal karyotyping was compared with an internally validated, laboratory-developed test based on next-generation sequencing in 212 Down syndrome and 1484 matched euploid pregnancies. None had been previously tested. Primary testing occurred at a CLIA-certified commercial laboratory, with cross validation by a CLIA-certified university laboratory.
Results: Down syndrome detection rate was 98.6% (209/212), the false-positive rate was 0.20% (3/1471), and the testing failed in 13 pregnancies (0.8%); all were euploid. Before unblinding, the primary testing laboratory also reported multiple alternative interpretations. Adjusting chromosome 21 counts for guanine cytosine base content had the largest impact on improving performance.
Conclusion: When applied to high-risk pregnancies, measuring maternal plasma DNA detects nearly all cases of Down syndrome at a very low false-positive rate. This method can substantially reduce the need for invasive diagnostic procedures and attendant procedure-related fetal losses. Although implementation issues need to be addressed, the evidence supports introducing this testing on a clinical basis.
Similar content being viewed by others
Main
Currently, the most effective prenatal screening tests for Down syndrome combine maternal age with information from sonographic measurement of nuchal translucency in the first trimester and measurements of several maternal serum screening markers obtained in the first and second trimesters.1,2 This approach detects up to 90% of all cases at a false-positive rate of 2%. Given the prevalence of Down syndrome, 1 of every 16 screen positive women offered invasive diagnostic testing (amniocentesis or chorionic villus sampling) will have an affected pregnancy and 15 will not. As many as 1 in 200 such invasive procedures are associated with fetal loss, a major adverse consequence of prenatal diagnosis.3,4 This has led to adjusting screening cutoffs to minimize the false-positive rate. In practice, false-positive rates of 5% are common.
The 1997 discovery that 3–6% of cell-free DNA in maternal blood was of fetal origin prompted studies to determine whether Down syndrome could be detected noninvasively.5 In 2008, two groups identified fetal Down syndrome, using massively parallel shotgun sequencing (MPSS).6,7 This technique sequences the first 36 bases of millions of DNA fragments to determine their specific chromosomal origin. If the fetus has a third chromosome 21, the percentage of chromosome 21 fragments is slightly higher than expected. Subsequent reports have extended these observations and suggest that a detection rate of at least 98% can be achieved at a false-positive rate of 2% or lower.8–10 Although promising, these studies were relatively small (range 13–86 Down syndrome cases and 34–410 euploid control samples), DNA sequencing was not performed in CLIA-certified laboratories, and throughput and turnaround times did not simulate clinical practice. The current independent, collaborative study addresses these and other shortcomings.
MATERIALS AND METHODS
See “Expanded Methods,” Appendix A, Supplemental Digital Content 1, http://links.lww.com/GIM/A213, for complete details.
Overall study design
Our study (clinicaltrials.gov NCT00877292) involved patients enrolled at 27 prenatal diagnostic centers worldwide (Enrollment Sites). Women at high risk for Down syndrome based on maternal age, family history or a positive serum and/or sonographic screening test provided consent, plasma samples, and demographic and pregnancy-related information. Institutional Review Board approval (or equivalent) was obtained at each site. Identification was by study code. Samples were drawn immediately before invasive testing, processed within 6 hours, stored at −80°C, and shipped on dry ice to the Coordinating Center. Within this cohort, we developed a nested case-control study, with blinded DNA testing for Down syndrome. We matched seven euploid samples to each case, based on gestational age (nearest week; same trimester), Enrollment Site, race (self-declared), and time in freezer (within 1 month). Assuming no false-negative results, 200 Down syndrome pregnancies (cases) had 80% power to reject 98% as the lower confidence interval (CI). The cases were distributed equally between first and second trimesters. For this study, we defined Down syndrome as 47, XY,+21 or 47, XX,+21; mosaics and twin pregnancies with Down syndrome were excluded.
Study coordination and sample storage were based at an independent academic medical center (Women & Infants Hospital). Frozen, coded samples (4 mL) were sent to the Sequenom Center for Molecular Medicine (SCMM, San Diego, CA) for testing. SCMM had no knowledge of the karyotype and simulated clinical testing, including quantifying turnaround time. A subset of samples was sent for testing at the Orphan Disease Testing Center at University of California at Los Angeles (UCLA; Los Angeles, CA), an independent academic laboratory experienced in DNA sequencing. Both laboratories were CLIA-certified, and both provided clinical interpretations using a standardized written protocol originally developed by SCMM.
Study integrity
We gave highest priority to ensuring integrity, reliability, and independence of this industry-funded study. We created a three-person Oversight Committee (see Acknowledgments), charged with assessing and providing recommendations on study design, conduct, analysis, and interpretation. The study protocol included Enrollment Site inspections, isolation of Enrollment Sites from the study sponsor, confirmatory testing by an independent academic laboratory, blinding of diagnostic test results on multiple levels, no remote computer access to outcome data, access to all raw data by the academic testing site, immediate file transfer of sequencing and interpretation results to the Coordinating Center, and use of file checksums to identify subsequent changes. SCMM provided the independent laboratory with similar equipment, training, interpretive software, and standard operating protocols.
The laboratory-developed test
MPSS has been described earlier.9 In brief, circulating cell-free DNA fragments are isolated from maternal plasma and quantified with an assay that determines the fetal contribution (fetal fraction).11 We used the remaining isolate to generate sequencing libraries, normalized and multiplexed to allow four samples to be run in a single flow cell lane (eight lanes per flow cell).9 We quantified DNA libraries using a microfluidics platform (Caliper Life Sciences, Hopkinton, MA) and generated clusters using the cBot platform (Illumina, Inc, San Diego, CA). We sequenced the flow cells on the Illumina HiSeq 2000 platform and analyzed resulting data using Illumina software. Computer interpretation provided a robust estimate of the SDs above or below the central estimate (z-score); z-scores at or above 3 were considered to be consistent with Down syndrome. The Director of the primary CLIA Laboratory (SCMM) reviewed results, initiated calls for testing second aliquots, and provided a final “signed out” interpretation for all pregnancies tested. The Director of the independent CLIA Laboratory (UCLA) did the same but without the ability to call for second sample aliquots. Each laboratory only had access to its own results.
Statistical analysis
The study would be paused if an interim analysis showed that more than 3 of 16 cases or 6 of 112 controls were misclassified. Although a matched study, we planned the analysis to be unmatched. We examined differences among groups and associations using χ2 test, t-test, analysis of variance (ANOVA), and linear regression (after appropriate transformations) using SAS (Cary, NC) and True Epistat (Richardson, TX). We computed CIs of proportions using the binomial distribution. P values were two-sided, and significance was at the 0.05 level.
RESULTS
Sample population
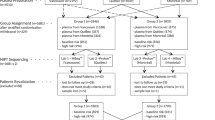
Between April 2009 and February 2011, 27 Enrollment Sites (Table 1) identified eligible pregnant women, obtained informed consent, and collected samples. Among 4664 enrollees, 218 singleton Down syndrome and 3930 singleton euploid pregnancies occurred. Figure 1 provides details on fetal outcomes, plasma sample status, and reasons why 279 women (6%) were excluded. None of the samples was included in previous publications or studies. A total of 4385 women (94%) had a singleton pregnancy, at least two suitable plasma samples and diagnostic test results. Of these, 97% were between 11 and 20 weeks' gestation, inclusive; 34% were in the first trimester. Similar numbers of Down syndrome fetuses (cases) were diagnosed in each trimester, and the first 212 enrolled were selected for testing. For each case, seven matched euploid pregnancies were chosen (1484). One control was later discovered to be trisomy 18 but was included as a “euploid” control.
Flow diagram displaying information about the enrolled patients and their pregnancies. Fetal karyotypes (or equivalent) were available for all but 51 enrolled women. For 116 women, the plasma samples were not considered adequate for testing (e.g., thawed during transit, more than 6 hours before being frozen, only one aliquot, and insufficient volume). An additional 112 women were excluded because of multiple gestations or existing fetal death. Among the remaining 4385 viable singleton pregnancies, 34% were obtained in the late first trimester and 66% in the early second trimester. A total of 212 Down syndrome cases were selected for testing, along with 1484 matched euploid controls (7:1). Among the 237 other outcomes were additional autosomal aneuploidies, sex chromosome aneuploidies, mosaics, and other chromosomal abnormalities.
Table 2 compares demographic and pregnancy-related information between cases and controls. Matching was successful. Median age was about 37 years in both groups; all were 18 years or older. Indications for diagnostic testing differed, with cases more likely to have an ultrasound abnormality or multiple indications. Samples were collected, processed, and frozen, on average, within 1 hour; all within 6 hours. Outcomes were based on karyotyping, except for two first trimester cases (quantitative polymerase chain reaction in one, and fluorescence in situ hybridization in the other, of products of conception after termination of a viable fetus with severe ultrasound abnormalities).
Fetal contribution to circulating free DNA
Before MPSS, extracted DNA was tested to determine the proportion of free DNA of fetal origin in maternal plasma (fetal fraction). Nearly all (1687/1696; 99.5%) had a final fetal fraction within acceptable limits (4–50%)9; the geometric mean was 13.4%. The lower cutoff was chosen to minimize false-negative results. The upper cutoff was chosen to alert the Laboratory Director that this represents a rare event. Nine had unacceptable levels; six below the threshold and three above. As the success of MPSS in identifying Down syndrome is highly dependent on the fetal fraction, 16 potential covariates (eFigs. B1–B16, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213) were explored (processing time, hemolysis, geographic region, indication for diagnostic testing, Enrollment Site, gestational age, maternal age, maternal weight, vaginal bleeding, maternal race, Caucasian ethnicity, fetal sex, freezer storage time, and effect of fetal fraction on DNA library concentration, number of matched sequences, and fetal outcome). A strong negative association of fetal fraction with maternal weight was observed in case and control women (eFig. B8, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213), with weights of 100, 150, and 250 pounds associated with predicted fetal fractions of 17.8%, 13.2%, and 7.3%, respectively. No association was found for gestational age, maternal race, or indication for testing. Other associations were small and usually nonsignificant.
Massively parallel shotgun sequencing testing for Down syndrome
Testing was performed over 9 weeks (January to March, 2011) by 30 scientists, molecular technicians/technologists with training on the assay protocols, and related instrumentation. Historical reference ranges were to be used for interpretation,9 with real-time review of new data a requirement. Review of the first few flow cells by the Laboratory Director (before sign out) revealed that adjustments to the reference data were necessary (Expanded Methods, Appendix A and eFigs. B17–B19, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213). After data from six flow cells were generated, results were assessed by the Oversight Committee according to the interim criteria, and the confidential decision was made to allow the testing to continue. At the conclusion of testing, but before unblinding, SCMM requested a second aliquot for 85 of the 90 test failures among the 1696 enrollees (5.3%; 95% CI, 4.3–6.5) (eFig. B36, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213). The second result was used for final interpretation.
Figure 2A shows the chromosome 21 z-score versus fetal fraction for all 212 Down syndrome and 1471 of 1484 euploid samples (excluding 13 failed samples). A strong positive association between fetal fraction and z-score existed for cases (after logarithmic transformation, slope = 0.676, P < 0.001) but not for controls (slope = 0.0022, P = 0.50). Four Down syndrome samples had z-scores below the cutoff of 3; all had fetal fractions of ≤7%. One of these had an initial z-score of 5.9 with one borderline quality failure; the repeat sample z-score was 2.9 (a borderline value consistent with the initial positive result). The Laboratory Director considered both results to make the interpretation. Therefore, signed out results (Fig. 2B) correctly identified 209 of 212 Down syndrome fetuses (detection rate of 98.6%; 95% CI, 95.9–99.7). Among the 1471 euploid samples, 3 had z-scores >3 over a range of fetal fractions and were incorrectly classified as Down syndrome, yielding a false-positive rate of 0.2% (95% CI, <0.1–0.6). For 13 women (13/1696 or 0.8%; 95% CI, 0.4–1.3), interpretation was not provided due to quality control failures on initial and repeat samples (six had fetal fractions <4%, one >50%), although their test results were available and usually “normal” (Fig. 2B). Laboratory results, sample handling, and pregnancy outcomes for the misclassified pregnancies were extensively checked for potential errors; none were identified (eTable B1, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213).
MPSS results for Down syndrome and matched euploid samples. A, The fetal fraction (x-axis) is shown versus the computer interpretation expressed in z-score (y-axis) for the 212 Down syndrome samples (large circles) and 1471 matched euploid samples (small circles). Not included in this figure are the 13 samples with repeated quality measure failures. The thin horizontal line is drawn at the z-score of 0, the approximate center of the euploid results, and shows that these results do not vary by fetal fraction. The dashed horizontal line at 3 indicates the cutoff level, above which the computer reports the result to be consistent with Down syndrome. Three euploid results fall above this cutoff level. The Down syndrome samples show a clear and significant positive relationship with fetal fraction; 208 of the samples are above the cutoff and four are below. All of those that fall below have relatively low fetal fractions (7%, 7%, 5%, and 4%). B, The clinical interpretation of all Down syndrome and euploid samples in the study. The interpretations are test positive for Down syndrome (DS+), test negative for Down syndrome (DS−), and test failure (Failure). Filled symbols indicate samples that have been tested twice, due to an inability to interpret the initial sample. Among the euploid pregnancies, 1468 were negative, 3 were positive, and 13 failed on the second aliquot as well. Among the Down syndrome pregnancies, 209 were positive and 3 were negative. One positive interpretation was associated with a z-score below 3 (2.9). The Laboratory Director combined this information from the repeated sample with a 5.9 score on the initial sample (with a borderline failure) to make the correct call. All other clinical interpretations agreed with the computer interpretation.
Analysis of the first 15 covariates versus z-score was performed (eFigs. B20–B34, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213). A strong negative association existed for maternal weight among cases; this association was weaker in controls. There was a small, but significant, positive association with gestational age in cases (eFig. B25, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213), with regressed z-scores at 11 and 19 weeks' gestation of 7.2 and 9.9, respectively. Other associations were small and usually nonsignificant.
Confirmation by an independent laboratory of testing performance
The UCLA laboratory performed cluster generation, DNA sequencing, and interpretation for a subset of 605 initial sample aliquots originally processed and tested by SCMM. This subset was randomly selected by the Coordinating Center from all complete groups of 92 patient samples (plates). Figure 3 shows a scatterplot of chromosome 21 z-scores for 578 samples successfully tested at both sites (96%). Correlations were high among both 77 Down syndrome and 501 euploid pregnancies (R = 0.80 and 0.83, respectively). Twenty-seven initial sample failures at one or both sites are not shown. In this subset of 578, the detection, false-positive, and initial failure rates for SCMM were 98.7%, 0.0%, and 4.4%, respectively. The corresponding rates for UCLA were 98.7%, 0.2%, and 3.9% (eTable B1, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213).
MPSS chromosome 21 test results from two laboratories in a subset of 605 samples. Computer-interpreted MPSS results are expressed as a z-score, with Sequenom Center for Molecular Medicine (SCMM) values on the x-axis and those from the UCLA laboratory on the y-axis. The figure shows the 77 Down syndrome and 501 euploid pregnancies that were successfully tested at both sites. The 27 samples that failed on the initial test at one or both sites are not included. The vertical and horizontal dotted lines show the z-score cutoff of 3. Among these samples, only one disagreement occurred. A euploid sample was misclassified by UCLA (z-score = 3.46) but correctly classified by SCMM (z-score = 2.02). Both groups misclassified one Down syndrome sample.
In another subset of 56 enrollees, duplicate 4 mL plasma samples were tested by each laboratory. One euploid sample failed at both sites (low fetal fraction). Two additional euploid samples failed sequencing at UCLA; their protocol did not allow retesting. Failure rates at SCMM and UCLA were 1.8% and 5.3%, respectively. Among 53 remaining samples, the two sites agreed on all quality parameters and interpretive results (eFig. B39, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213). At both laboratories, the detection and false-positive rates were 100% and 0%, respectively.
Post hoc analysis
The large sample size provided an opportunity to investigate alternative methods of interpreting the MPSS results. After sign out, but before laboratory unblinding, chromosome 21 percent results were adjusted by the SCMM laboratory for GC content, a process shown to improve MPSS performance,12,13 as well as filtered with respect to The Repeat Mask (www.repeatmasker.org/PreMaskedGenomes.html) and the results forwarded to the Coordinating Center to determine whether alternative interpretive algorithms might perform better, be more robust, or both. Analysis showed that control results varied by flow cell or by plate (three flow cells that are batch processed) (ANOVA, F = 13.5, P < 0.001), but the SD was constant (ANOVA, F = 1.2, P = 0.23), allowing us to convert the GC-adjusted results to multiples of the plate median. Multiples of the plate median values in Down syndrome and euploid pregnancies were completely separate, except for the one persistent false-negative result (eFig. B41, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213). Adjusting flow-cell specific z-scores also improved performance, with two false negative and one false positive result remaining (eFig. B42, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213). None of these post hoc analyses was available at the time the clinical interpretation was made.
Clinical implications
Two thousand one hundred and sixteen initial patient samples (1696 reported here and 420 other patient samples) were tested with a throughput of 235 patients per week using two HiSeq 2000 platforms. Turnaround time (sample thaw to sign out) improved over the 9 weeks of testing, meeting a 10-day target for 18 of the final 20 flow cells (eFig. B35, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213). This does not include the 5% of samples that required a second aliquot, although turnaround time for these would not double because failures are often discovered early in the testing process.
To assess utility, a simple model (eFig. B39, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213) compares current diagnostic protocols for Down syndrome with one that inserts MPSS between identification of high-risk pregnancy and invasive diagnosis. Assume 100,000 women at high risk for Down syndrome, with one affected pregnancy for every 32 normal pregnancies, diagnostic testing costs of $1,000 per patient (eFig. B39, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213), and a procedure-related fetal loss rate of 1 in 200.3,4,14 Complete uptake of invasive testing by high-risk women would detect 3,000 cases at a cost of $100 million and 500 procedure-related losses. Complete uptake of MPSS testing by all high-risk women, followed by invasive testing in those with positive MPSS results (along with those who failed testing), would detect 2,958 cases (42 missed) at a cost of $3.9 million and 20 losses. The difference in financial costs for the two protocols could help offset MPSS testing costs. Assigning a dollar value to the 480 potentially avoidable procedure-related losses is difficult, but they are an equally important consideration. If the procedure-related loss rate were lower than 1 in 200, the absolute number of losses would decrease, but the proportional reduction would remain the same.
DISCUSSION
This study extends the findings of previously published reports.8–10 Together with our report a total of 350 Down syndrome and 2061 control pregnancies have been reported and document 99.0% sensitivity and specificity (95% CI, 98.2–99.8%, I2 = 0%; eTable B3, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213), providing definitive evidence of the clinical validity of a test for Down syndrome based on MPSS. A positive test result increased Down syndrome risk by 490-fold (98.6% detection/0.2% false-positive rate); a negative result reduced risk by 72-fold (99.8%/1.4%). Testing was successful in 992 of every 1000 women. Although 5.3% of initial tests failed quality checks, 82% of these were resolved after testing second aliquots. Most remaining test failures were associated with a low fetal fraction, which might be solved by repeat sampling a week or two later in pregnancy. MPSS performance was confirmed by the independent laboratory (Fig. 3; eTable B3, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213) using original plasma samples and plasma DNA preparations.
The current study handled large numbers of samples (collection, processing, freezing, and shipping) by 27 Enrollment Sites; simulating expected clinical practice. Our findings support MPSS performance across a broad gestational age range, among various racial/ethnic groups, for all maternal ages and for all diagnostic testing indications (eFig. B23, Appendix B, Supplemental Digital Content 1, http://links.lww.com/GIM/A213). Performance is not affected by vaginal bleeding or sample hemolysis and is robust to sample processing time up to 6 hours. Because of the well-described dilution effect of increased blood volume,15 test failures are more common in heavier women. Accounting for fetal fraction in the interpretation may be warranted. Overall, most women with false-positive screening results will avoid invasive testing, while nearly all affected pregnancies will be confidently diagnosed by conventional invasive means. The present study supports offering MPSS to women identified as being at high risk for Down syndrome, taking into account the test's complexity and resources required. Were testing to occur at least twice a week, the turnaround time for 95% of patient results would be comparable with that currently available for cytogenetic analysis of amniotic fluid cells and chorionic villus sampling. Availability of MPSS could also justify lowering serum/ultrasound screening cutoffs, resulting in higher Down syndrome detection. This study documents, for the first time, an inherent variability from flow-cell to flow-cell. Accounting for these changes improves clinical performance. How best to perform such adjustments needs more study.
Post hoc analyses resulted in reduced false-negative and false-positive results, mostly because of adjustments for GC content. This constitutes strong evidence that MPSS performance will be better when testing is introduced into practice. This study also provides evidence that MPSS can be translated from research to a clinical setting with reasonable turnaround and throughput. Certain implementation issues deserve attention. A collection tube that allows storage and shipment at ambient temperature without affecting cell-free DNA levels16 would be helpful. Currently, samples must be processed, frozen, and shipped on dry ice, similar to the protocol followed in our study. As this was an observational study, a demonstration project showing efficacy in clinical settings is warranted. Educational materials for both patients and providers need to be developed and validated to help ensure informed decision making. Additional concerns include reimbursement and development of relevant professional guidelines. Some have suggested that testing fetal DNA raises new ethical questions.17–19 In the recommended setting of MPSS testing of women at high risk, many of these questions are not relevant.
A major goal in the field of prenatal screening has been to reduce the need for invasive procedures.20 MPSS testing cannot yet be considered diagnostic. However, offering MPSS testing to women already at high risk for Down syndrome can reduce procedure-related losses by up to 96%, while maintaining high detection. Confirmation by invasive testing is still needed. This study, along with previous reports, documents high performance, but we extend the evidence by performing the testing in a CLIA-certified laboratory, having second aliquots available for initial failures, monitoring turnaround time, assessing operator to operator and machine to machine variability, validating a subset of sample results in an independent academic clinical laboratory, and integrating a medical geneticist/laboratory director into the reporting process. This report does not address other chromosome abnormalities13 or events such as twin pregnancies. As the technology moves forward, such refinements will become available. Although some implementation issues still need to be addressed, the evidence warrants introduction of this test on a clinical basis to women at high risk of Down syndrome, before invasive diagnostic testing.
REFERENCES
Malone FD, Canick JA, Ball RH, et al. First-trimester or second-trimester screening, or both, for Down's syndrome. N Engl J Med 2005; 353: 2001–2011.
Wald NJ, Watt HC, Hackshaw AK . Integrated screening for Down's syndrome on the basis of tests performed during the first and second trimesters. N Engl J Med 1999; 341: 461–467.
ACOG. ACOG Practice Bulletin No. 88, December 2007. Invasive prenatal testing for aneuploidy. Obstet Gynecol 2007; 110: 1459–1467.
Tabor A, Philip J, Madsen M, Bang J, Obel EB, Nørgaard-Pedersen, Randomised controlled trial of genetic amniocentesis in 4606 low-risk women. Lancet 1986; 1: 1287–1293.
Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet 1997; 350: 485–487.
Chiu RW, Chan KC, Gao Y, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci USA 2008; 105: 20458–20463.
Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR . Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA 2008; 105: 16266–16271.
Chiu RW, Akolekar R, Zheng YW, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ 2011; 342: c7401.
Ehrich M, Deciu C, Zwiefelhofer T, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am J Obstet Gynecol 2011; 204: 205.e201–205.e211.
Sehnert AJ, Rhees B, Comstock D, et al. Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell-free fetal DNA from maternal blood. Clin Chem 2011; 57: 1042–1049.
Nygren AO, Dean J, Jensen TJ, et al. Quantification of fetal DNA by use of methylation-based DNA discrimination. Clin Chem 2010; 56: 1627–1635.
Alkan C, Kidd JM, Marques-Bonet T, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet 2009; 41: 1061–1067.
Chen EZ, Chiu RWK, Sun H, Akolekar R, et al. Noninvasive prenatal diagnosis of fetal trisomy 18 and trisomy 13 by maternal plasma DNA sequencing. PLoS One 2011; 6: e21791.
Tabor A, Alfirevic Z . Update on procedure-related risks for prenatal diagnosis techniques. Fetal Diagn Ther 2010; 27: 1–7.
Neveux LM, Palomaki GE, Larrivee DA, Knight GJ, Haddow JE . Refinements in managing maternal weight adjustment for interpreting prenatal screening results. Prenat Diagn 1996; 16: 1115–1119.
Fernando MR, Chen K, Norton S, et al. A new methodology to preserve the original proportion and integrity of cell-free fetal DNA in maternal plasma during sample processing and storage. Prenat Diagn 2010; 30: 418–424.
Benn PA, Chapman AR . Ethical challenges in providing noninvasive prenatal diagnosis. Curr Opin Obstet Gynecol 2010; 22: 128–134.
Deans Z, Newson AJ . Should non-invasiveness change informed consent procedures for prenatal diagnosis?. Health Care Anal 2011; 19: 122–132.
Newson AJ . Ethical aspects arising from non-invasive fetal diagnosis. Semin Fetal Neonatal Med 2008; 13: 103–108.
Haddow JE, Palomaki GE, Knight GJ, et al. Reducing the need for amniocentesis in women 35 years of age or older with serum markers for screening. N Engl J Med 1994; 330: 1114–1118.
Acknowledgements
Sequenom, Inc. fully funded the project through a grant to Women & Infants Hospital of Rhode Island.
We thank the Oversight Committee members Michael T. Mennuti, MD (Chair), University of Pennsylvania Medical Center, Philadelphia, Pennsylvania; George A. Macones, MD (Member), MSCE, Washington University School of Medicine, St. Louis, Missouri; Iris Schrijver, MD (Member), Stanford University and Stanford School of Medicine, Stanford, California; and Wayne W. Grody, MD, PhD (Past Member), David Geffen School of Medicine at UCLA, Los Angeles, California, for their independent assessment of the study design and conduct and their thoughtful advice throughout the planning and active enrollment phases. Member activities were compensated with study funds.
We thank the Enrollment Site personnel for their study-funded participation in the study. Those participants include North York General Hospital, Genetics Program, Toronto, Ontario, Canada: Wendy S. Meschino, MD, and Clare Gibbons, MS; Istituto G. Gaslini, Genova, Italy: Pierangela De Biasio, MD, Lucia Vaccari, MD, Sabrina Bertoldi, MD, Daniela Pastorino MD, Margherita Giordano, MD, Gianluca Caridi, BCS, and Monica Dagnino, BCS; Hospital Clinic Barcelona, Barcelona, Catalonia, Spain: Antoni Borrell, MD, PhD, Agusti Seres, MD, Roser Febrero, BSc, Iratxe Torre, BSc, and Maribel Grande, PhD; Centrum Lekarske Genetiky, Ceske Budejovice, Czech Republic: David Cutka, MD, and Karel Cutka, MD; Hospital Italiano, Buenos Aires, Argentina: Lucas Otaño, MD, PhD, Gustavo Izbizky, MD, and Carla Sesarini, MS; IWK Health Center, Dalhousie University, Halifax, Canada: Michiel Van den Hof, MD, Jo-Ann Brock, MD, PhD, and Cora Fanning, BN, RN; Royal College of Surgeons in Ireland, Rotunda Hospital, Dublin, Ireland: Fergal Malone, MD, Etaoin Kent, MD, Julia Unterscheider, MD, and Grainne McSorley, RGN; First Department of OBGYN, Semmelweis, University, Budapest, Hungary: Csaba Papp, MD, PhD, and Balint Nagy, PhD; IMALAB s.r.o. Medical Laboratories and Prediko s.r.o. Genetic and Prenatal Screening Centre, Zlin, Czech Republic: Jaroslav Loucky, RNDr, and Petr Polak, MD; Genetics Section, Centro de Educacion Medica e Investigaciones Clinicas (CEMIC), Buenos Aires, Argentina: Laura Igarzabal, MD, and Florencia Petracchi, MD; The University of Iowa Hospitals and Clinics, Department of Obstetrics and Gynecology, Iowa City, Iowa: Kristi Borowski, MD, Roger Williamson, MD, Diedre Fleener, RN, and Bruce Bedell, MD; Prenatal Diagnosis Center, Women & Infants Hospital, Providence, Rhode Island: Barbara M O'Brien, MD, and Jacquelyn Halliday, MS; University of Pécs, Clinical Center Department of Obstetrics and Gynecology, Pécs, Hungary: Béla Veszprémi, MD, PhD, and Gabor Molnar, MD; The Center for Women's Reproductive Health at the University of Alabama at Birmingham, Birmingham, Alabama: Joseph Biggio, MD, and Rachel Copper, MSN, WHNP-BC; Rambam Medical Center, Haifa, Israel: Zeev Weiner, MD; Cedars Sinai Prenatal Diagnostic Center, Division of Maternal-Fetal Medicine, Los Angeles, California: John Williams, MD, Deebra Smith, MPH, and Rennatha Edwards; Northwestern University Feinberg School of Medicine, Chicago, Illinois: Jeffrey Dungan, MD, William Grobman, MD, Lee P. Shulman, MD, and Suzanne Banuvar, MPA/HSA; Henry Ford Hospital, Department of Genetics, Detroit, Michigan: Jacquelyn Roberson, MD, and Ellen Martindell, RN, BSN; Department of Obstetrics and Gynecology, University of Virginia, Charlottesville, Virginia: Devereux N. Saller, Jr, MS, MD, and Karen A. Ventura, MS; Children's and Women's Health Centre of British Columbia, Vancouver, British Columbia, Canada: Sylvie Langlois, MD, FRCPC, and Patricia Power, MSc; Intermountain Health care and ARUP Laboratories, Salt Lake City, Utah: Nancy Rose, MD, Elaine Lyon, PhD, and Danielle LaGrave, MS, LCGC; Brigham and Women's Hospital, Boston, Massachusetts: Louise Wilkins-Haug, MD, and Diane Ahern, MS, CGC; Baylor College of Medicine, Department of Obstetrics and Gynecology, Houston, Texas: Anthony Johnson, DO, Blair Stevens, MS, Ping Fang, PhD, and David Vo; Department of Genetics, Yale University School of Medicine, New Haven, Connecticut: Maurice J. Mahoney, MD, JD, Joanne R. Florio, MS, and Peining Li, PhD; New Beginnings Perinatal Consultants: Providence, Rhode Island: Marshall Carpenter, MD; University of Calgary, Early Assessment Risk Program, Calgary, Alberta, Canada: Jo-Ann Johnson, MD, and Melanie Pastuck, RN; University of Sydney, Perinatal Research Group, Kolling Institute of Medical Research, Sydney, New South Wales, Australia: Vitomir Tasevski, PhD, and Jonathan Morris, MD, PhD.
We thank the study-funded Coordinating Center staff at Women & Infants Hospital, Providence, Rhode Island, and in Standish Maine; specifically, George J. Knight, PhD, for assistance in protocol development and oversight, Regina Traficante, PhD, for her role as Rhode Island study coordinator, Cheryl Felber for technical assistance, and Joanne Beaudoin for administrative assistance. We also thank Cindy Steinort (Electric Dreams, Inc, Scarborough, ME) for developing the study-specific data management system.
We thank Sequenom Center for Molecular Medicine staff, including Roger Tim, Vivian Lu, John Tynan, Lesley Cagasan, Tricia Zwiefelhofer, Ron McCullough, Erin McCarthy, Anders Nygren, Jarrod Dean, Taylor Jensen, Frank Aquino, Sonia Espina-Grenie, Mengjia Tang, Julia Clemens, Wen Wu, Daniela Hubbard, Yabo Jin, Toni Paladino, Anna Maria Al-Khouri, Edlyn Mendoza, Paul Oeth, Kyle Ingersoll, Nick Miltgen, Anna Van Agtmael, Molly Dobb, Carlee Koessel, Shirin Fitzgerald, Lyle Rawlings, Melissa O'Day, Stephanie Lin, Haiping Lu, Rochelle Jean-Jacques, Vach Angkachatchai, Lin Tang, Tim Lu, Zhanyang Zhu, Zeljko Dzakula, Christine Chen, Tom Wang, Ryan Biltz, Benjamin Najara, Jennifer Wolanski, and David A. Henderson for technical execution of the study.
We thank UCLA staff, including Zugen Chen for supervision of the next generation resource, Traci Toy for the project coordination and assay performance, Kingshuk Das for clinical interpretation, and Bret Harry for software implementation and processing of sample data.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure: Palomaki and Canick (Co-Principal Investigators) were members of the Sequenom Clinical Advisory Board for 6 months and resigned when the study was funded. Van den Boom, Ehrich, Bombard, and Deciu are employees and shareholders of Sequenom, Inc.
Role of the Sponsor: Sequenom Center for Molecular Medicine (SCMM) was responsible for developing an internally validated laboratory developed test (LDT) for detecting Down syndrome in maternal plasma using MPSS and for providing clinical interpretation of the test results. SCMM also identified, equipped, and trained an independent laboratory to test a subset of samples through a separate contract with UCLA. The sponsor did not control study design, identify, or communicate with Enrollment Sites, thaw or test samples prior to the formal testing period, have access to patient information prior to all testing being completed, analyze study results, prepare drafts of the manuscript, or have final decisions on manuscript content.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.geneticsinmedicine.org).
Rights and permissions
About this article
Cite this article
Palomaki, G., Kloza, E., Lambert-Messerlian, G. et al. DNA sequencing of maternal plasma to detect Down syndrome: An international clinical validation study. Genet Med 13, 913–920 (2011). https://doi.org/10.1097/GIM.0b013e3182368a0e
Published:
Issue Date:
DOI: https://doi.org/10.1097/GIM.0b013e3182368a0e
Keywords
This article is cited by
-
Laboratory performance of genome-wide cfDNA for copy number variants as compared to prenatal microarray
Molecular Cytogenetics (2023)
-
Estimation of cell-free fetal DNA fraction from maternal plasma based on linkage disequilibrium information
npj Genomic Medicine (2021)
-
Noninvasive prenatal testing: from aneuploidy to single genes
Human Genetics (2020)
-
Recent Advances in the Noninvasive Prenatal Testing for Chromosomal Abnormalities Using Maternal Plasma DNA
Journal of Fetal Medicine (2020)
-
The consequences of implementing non-invasive prenatal testing with cell-free foetal DNA for the detection of Down syndrome in the Spanish National Health Service: a cost-effectiveness analysis
Cost Effectiveness and Resource Allocation (2019)