Abstract
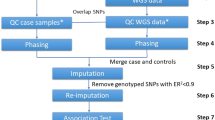
The Affymetrix GeneChip Human Mapping 500K array is common for genome-wide association studies (GWASs). Recent findings highlight the importance of accurate genotype calling algorithms to reduce the inflation in Type I and Type II error rates. Differential results due to genotype calling errors can introduce severe bias in case–control association study results. Using data from the Wellcome Trust Case Control Consortium, 1991 individuals with coronary artery disease (CAD) and 1500 controls from the UK Blood Services (NBS) were genotyped on the Affymetrix 500K array. Different batch sizes and compositions were used in the Bayesian Robust Linear Model with Mahalanobis distance classifier (BRLMM) genotype calling algorithm to assess the batch effect on downstream association analysis. Results show that composition (cases and controls genotyped simultaneously or separate) and size (number of individuals processed by BRLMM at a time) can create 2–3% discordance in the results for quality control and statistical analysis and may contribute to the lack of reproducibility between GWASs. The changes in batch size are largely responsible for differential single-nucleotide polymorphism results, yet we observe evidence of an interactive effect of batch size and composition that contributes to discordant results in the list of significantly associated loci.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Kingsmore SF, Lindquist IE, Mudge J, Gessler DD, Beavis WD . Genome-wide association studies: progress and potential for drug discovery and development. Nat Rev Drug Discov 2008; 7: 221–230.
Donnelly P . Progress and challenges in genome-wide association studies in humans. Nature 2008; 456: 728–731.
Clayton DG, Walker NM, Smyth DJ, Pask R, Cooper JD, Maier LM et al. Population structure, differential bias and genomic control in large-scale, case–control association study. Nat Genet 2008; 37: 1243–1246.
Di X, Matsuzaki H, Webster TA, Hubbell E, Liu G, Dong S et al. Dynamic model based algorithms for screening and genotyping over 100k SNPs on oligonucleotide microarrrays. Bioinformatics 2005; 21: 1958–1963.
Carvalho B, Bengtsson H, Speed TP, Irizarry RA . Exploration, normalization, and genotype calls of high-density oligonucleotide snp array data. Biostatistics 2007; 8: 485–499.
Lin S, Carvalho B, Cutler DJ, Arking DE, Chakravarti A, Irizarry RA . Validation and extension of an empirical bayes method for snp calling on affymetrix microarrays. Genome Biol 2008; 9: R63.
The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 case of seven common diseases and 3,000 shared controls. Nature 2007; 447: 661–678.
Winkelmann J, Schormair B, Lichtner P, Ripke S, Xiong L, Jalilizadeh S et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet 2007; 39: 1000–1006.
Meisinger C, Prokisch H, Gieger C, Soranzo N, Mehta D, Rosskopf D et al. A genome-wide association study identifies three loci associated with mean platelet volume. Am J Hum Genet 2008; 84: 66–71.
Gold B, Kirchhoff T, Stefanov S, Lautenberger J, Viale A, Garber J et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci USA 2008; 105: 4340–4345.
Affymetrix White Paper Publication. BRLMM: an improved genotype calling method for the genechip human mapping 500k array set http://www.affymetrix.com/support/technical/whitepapers/brlmmwhitepaper.pdf.
Plagnol V, Cooper JD, Todd JA, Clayton DG . A method to address differential bias in genotyping in large-scale association studies. PLoS Genet 2007; 3: 759–767.
Hong H, Su Z, Ge W, Shi L, Perkins R, Fang H et al. Assessing batch effect of genotype calling algorithm brlmm for affymetrix genechip human mapping 500k array set using 270 hapmap samples. BMC Bioinformatics 2008; 9 (Suppl 9): S17 .
Miyagawa T, Nishida N, Ohashi J, Kimura R, Fujimoto A, Kawashima M et al. Appropriate data cleaning methods for genome-wide association study. J Hum Genet 2008; 53: 886–893.
Anney RJ, Kenny E, O’Dushlaine CT, Lasky-Su J, Franke B, Morris DW et al. Non-random error in genotype calling procedures: Implications for family-based and case-control genome-wide association studies. Am J Med Genet B (Neuropsychiatr Genet) 2008; 147: 1379–1386.
Carvalho BS, Louis TA, Irizarry RA . Quantifying uncertainty in genotype calls. Bioinformatics 2010; 26: 242–249.
MicroArray Quality Control Consortium. The microarray quality control (maqc) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol 2006; 24: 1151–1161.
Acknowledgements
We thank all members of the GWAWG and MAQC for their contribution to this study. We also thank the members of the WTCCC for providing access to the data and the anonymous reviewers, whose comments and insight has made this a much more effective paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Disclaimer
The views presented in this article do not necessarily reflect those of the US Food and Drug Administration.
Rights and permissions
About this article
Cite this article
Miclaus, K., Wolfinger, R., Vega, S. et al. Batch effects in the BRLMM genotype calling algorithm influence GWAS results for the Affymetrix 500K array. Pharmacogenomics J 10, 336–346 (2010). https://doi.org/10.1038/tpj.2010.36
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tpj.2010.36
Keywords
This article is cited by
-
Genotype calling of triploid offspring from diploid parents
Genetics Selection Evolution (2020)
-
Systematic review of genetic polymorphisms associated with psychoneurological symptoms in breast cancer survivors
Supportive Care in Cancer (2019)
-
SNP genotype calling and quality control for multi-batch-based studies
Genes & Genomics (2019)
-
Identifying and mitigating batch effects in whole genome sequencing data
BMC Bioinformatics (2017)
-
Variability in GWAS analysis: the impact of genotype calling algorithm inconsistencies
The Pharmacogenomics Journal (2010)