Abstract
Nitric oxide (NO) signaling controls various metabolic pathways in bacteria and higher eukaryotes. Cellular enzymes synthesize and detoxify NO; however, a mechanism that controls its cellular homeostasis has not been identified. Here, we found a nitrogen oxide cycle involving nitrate reductase (Nar) and the NO dioxygenase flavohemoglobin (Fhb), that facilitate inter-conversion of nitrate, nitrite and NO in the actinobacterium Streptomyces coelicolor. This cycle regulates cellular NO levels, bacterial antibiotic production and morphological differentiation. NO down-regulates Nar and up-regulates Fhb gene expression via the NO-dependent transcriptional factors DevSR and NsrR, respectively, which are involved in the auto-regulation mechanism of intracellular NO levels. Nitrite generated by the NO cycles induces gene expression in neighboring cells, indicating an additional role of the cycle as a producer of a transmittable inter-cellular communication molecule.
Similar content being viewed by others
Introduction
Nitric oxide (NO) is a freely diffusible neutral gas that acts as an important signaling molecule to control metabolic pathways in bacteria and higher eukaryotes. Since NO is highly reactive and toxic for living cells, cells must have strict control over intracellular NO levels. The genus Streptomyces includes bacteria that produce many commercially useful secondary metabolites that are extremely important to humans. They follow an elaborate life cycle that includes vegetative (or substrate) mycelial growth, aerial mycelial growth and sporulation. Recent studies suggest that actinobacteria require NO to regulate various metabolic pathways1,2,3,4,5, however, little is known about the mechanism by which actinobacteria generate NO, except for NO synthase (NOS) distribution in a limited number of actinobacterial species.
Recently, we reported a unique nitrogen metabolism in Streptomyces antibioticus, in which organic nitrogen was mineralized to form nitrogen oxide species, nitrite (NO2−), nitrate (NO3−) and NO during vegetative cell growth under aerobic conditions and NO2− was excreted into the medium6. Since arginine analogs inhibited the production of NO2− and NO, we suggested that NOS is involved in the NO2− -formation and proposed a NO2− -forming pathway (pathway 1, below), although presence of a NOS enzyme has not been demonstrated in S. antibioticus.

We also showed that S. antibioticus produces a large amount of flavohemoglobin (Fhb) and oxidizes NO to generate nitrate (NO3−)7. The bacterium produces Fhb without exogenous nitrosative stress under NO2−-producing conditions, which is in contrast to most bacteria that produce Fhb only in the presence of exogenous NO8. Membrane-bound NO3− reductase (Nar) catalyzes the last step in the NO2−-forming pathway. Some bacteria produce Nar in the presence of NO3− for anaerobic respiration (denitrification)9, while S. antibioticus produces Nar and NO2− in the absence of exogenous NO3− under aerobic conditions6,10. Thus, both Fhb and Nar are produced in S. antibioticus to form NO2− under conditions that are different from those known in many other bacteria. Given that production of NO2− synchronized with rapid cell growth and was inhibited by glucose or glycerol, we suggested that the “NO2−-forming pathway” is an energy-producing metabolic reaction, although this conclusion requires further investigation.
This study investigated NO2− production by a model actinobacterium, S. coelicolor A3(2). Although S. coelicolor A3(2) produces and excretes NO2− like S. antibioticus, the NO-producing mechanism is likely to be different from the latter since the S. coelicolor genome11 does not encode a gene for NOS. In addition, no gene encoding the dissimilatory, NO-generating nitrite reductase was found in the genome. Here we demonstrated a NO-formation via NO2− produced from organic nitrogen and homeostatic regulation of cellular NO in S. coelicolor. The endogenously formed NO is controlled by the nitrogen oxide cycle and acts as a signaling molecule for antibiotic production and morphological differentiation.
Results
S. coelicolor nitrogen-oxide cycle tunes endogenous NO concentration
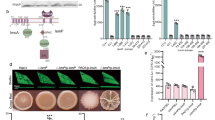
We found that S. coelicolor A3 (2) M145 (M145) excreted NO2− into the medium when cultured with organic nitrogen as the nitrogen source under NO2−-producing conditions6 (Fig. 1a). The NO2− _production was also observed when M145 was grown on minimal medium containing L-asparagine as the sole nitrogen source (chemically defined medium) (supplementary Fig. 1), showing that M145 can convert organic nitrogen to NO2−. NO2− concentration increased for 72 h, indicating that S. coelicolor produces NO2− from organic nitrogen during the early vegetative growth phase. The S. coelicolor A3(2) has three Nar enzyme homologs (Nar1, Nar2 and Nar3 encoded by narGHJI, narG2H2J2I2 and narG3H3J3I3, respectively). We constructed mutants lacking combinations of the three narG paralogs, each of which encodes the catalytic subunits of Nar. (∆narG, ∆narG2, ∆narG3, ∆narG2/G3 and ∆narG/G2/G3). The wild-type M145 strain cultured under the NO2−-producing conditions showed Nar activity in cell membrane fractions (Fig. 1b). The ∆narG2/G3 and ∆narG/G2/G3 mutants showed drastic decrease in this activity (Fig. 1b) accompanied by decreased NO2− production (Fig. 1c). The ∆narG/G2/G3 mutant excreted NO3− instead of NO2− (Fig. 1a), indicating that Nar reduces NO3− to NO2−. Introducing the narG2H2J2I2 operon into the ∆narG/G2/G3 mutant restored the NO2−-production (Fig. 1c). ∆narG mutant produced almost the same level of Nar activity as the wild type M145 strain (Fig. 1c). The result indicated that narG2 and narG3 but not narG are responsible for NO2− formation from organic nitrogen in the culture medium. Recently, Fischer et al. proposed that all three Nars are synthesized in S. coelicolor during the aerobic growth independent of the presence of NO3− and they further indicated that narG is mainly working in spore, narG2 and narG3 are mainly working in mycelium, respectively. Thus, present results of narG2 and narG3 expression correspond with the results reported by Fischer et al.12,13.
Nitrite and nitrate production by S. coelicolor A3(2) M145 and its derivatives.
(a) Spores of S. coelicolor A3(2) M145 or its defective strains were inoculated on cellophane-covered YEME-gln plates at intervals of 1 cm (a total of 45 spots). Levels of NO2− and NO3− extracted from the medium plate at each cultivation time as described in Materials and Methods were determined. Open circle, NO2− production by M145; closed circle, NO3− by M145; open square, NO2− by ∆narG/G2/G3; closed square, NO3− by ∆narG/G2/G3. (b) Nar activity in the membrane fraction of M145 and its derivatives. Cells were cultivated as described in (a) for 48 h and the membrane fraction was prepared as described in METHODS. (c) Each strain was cultivated as in (a) except that spores were inoculated directly on the agar plate (without cellophane). Photographs of each strain: upper line, grown cells without treatment; lower line, NO2− production was visualized by dyeing with 1 ml Griess reagent. Error bars indicate standard deviation (n = 3).
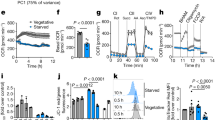
Since NO2− as well as NO3− are produced as endogenously oxidative metabolites of NO, we investigated NO accumulation by using a fluorometric method and observed that NO production activity was present in M145 (Fig. 2a). The gene knockout of hmpA (SCO7472, encodes FhbA) (∆hmpA) accumulated more NO than did the M145 strain (Fig. 2a) and complementation with the hmpA gene (∆hmpA:: hmpA) attenuated NO accumulation to the same level as that in M145 (Fig. 2a). Western blot analysis detected the hmpA gene product (flavohemoglobin, Fhb) in the NO2−-producing cells (Fig. 2b), indicating that the NO dioxygenase activity of Fhb8 led to lowered intracellular NO levels.
Production of endogenous NO and NO dioxygenase (Fhb) in S. coelicolor.
(a) Spores of each strain were inoculated at the center of a YEME-gln medium plate (one spot, without cellophane) and cells were grown for the indicated period. Cells were then collected with a toothpick and treated with 10 μM DAF-2DA as described previously. NO2− (50 μM) was added into the growth medium (bottom figure). hotographs under visible light (L) or fluorescence (F) were taken using FLUOVIEW FV300 System. Scale bar, 100 μm (b) Soluble proteins (7 μg) from each strain grown for the indicated period were used to detect Fhb production by western blotting. NO2− (50 μM) was added into the growth medium (bottom figure). (c) Production of FhbA is controlled by the negative transcriptional factor NsrR. Cells were cultivated as in (a) for 48 h. Soluble proteins (7 μg) of each strain were prepared as in (b).
When grown under the NO2−-producing conditions, the ∆narG/G2/G3 cells accumulated little NO, whereas exogenous addition of NO2− to the cells restored the NO formation (Fig. 2a). This showed that NO was formed from NO2− and that the ∆narG/G2/G3 cells could not form NO because they could not produce NO2− (Fig. 1a,c). The ∆narG/G2/G3 cells produced little Fhb and addition of an NO donor (50 μM NO2−) to the medium restored its production (Fig. 2b). Transcription of hmp is known to be negatively regulated by NsrR14,15, which loses the ability to repress hmp upon exposure to NO. Disruption of nsrR in the ∆narG/G2/G3 cells also recovered the defect of FhbA production (Fig. 2c). These results are consistent with the notion that NO is formed from NO2− that is produced by Nar and derepresses Fhb production through NsrR transcription control. The present results demonstrate that Nar encoded by narG2 and narG3 reduces NO3− to NO2−, which is converted to NO, while Fhb oxidizes NO to NO3−. These enzymes constitute the metabolic cycle of nitrogen oxides, which can participate in the cellular processes without exogenous supply of either of the nitrogen oxides.
The expression of FhbA was observed in ∆narG2/G3 mutant only at early stage of growth (24 h) (Fig. 2b). This suggests that narG supported the expression of FhbA, confirming that narG acts during at spore germination stage and also confirms that narG2 and narG3 are working during vegetative growth phase to supply NO via NO2−.
Endogenous NO promotes antibiotic production and regulates differentiation
Addition of NO donors to S. coelicolor cultures up-regulated transcription of redD, which encodes a positive regulator of undecylprodigiosin (Red) synthesis genes16 and Red production (Fig. 3a). Direct supply of NO by NOC5 induced redD transcription more effectively than NO2− (Fig. 3a, left), which indicates that NO directly regulates redD transcription. The ∆hmpA strain produced more Red than did the M145 strain (Fig. 3b) whereas ∆narG2 and ∆narG/G2/G3 mutants produced little Red (Fig. 3c) in accordance with the roles of Fhb and Nar in sequestering and producing NO, respectively. Addtion of NO donor (100 μM NO2−) restored the Red production by the ∆narG/G2/G3 mutant (Fig. 3c), showing that NO2− production from NO3− by Nar is necessary for NO formation. Higher concentration of NO2− increased Red production by the M145 and ∆narG/G2/G3 strains, but redD gene knockout confirms no NO-dependent Red production (Fig. 3a, right). These results indicated that the bacterial Nar and FhbA control cellular NO levels and regulate Red production. Moreover, the ∆narG/G2/G3 mutant developed aerial mycelia after cultivation for 120 h, which is much earlier than differentiation of aerial mycelia in M145. Addition of 100 μM NO2− to the medium recovered this phenotype (Fig. 3c), indicating that endogenously generated NO delays aerial mycelia development.
NO derived from NO2− regulates antibiotic (RED) production and morphological differentiation in S. coelicolor.
Cells were grown for the indicated period as in Fig. 2 except in panel (a). Scale bar, 1 mm. (a) NO2−- or NO-dependent redD expression (left panel) or RED production (right panel). Cells were grown for 48 h as in Fig. 1a and cells on the cellophane membrane were transferred to a fresh medium plate and further incubated. Left panel, cells were incubated at 30 °C for 60 (NO2−) or 25 (NOC5) min, total RNA was extracted and expression was determined by qPCR. Right panel, effects of NO2− on RED production, calculated as μmol RED/g wet weight cells. White bar, wild M145; dark bar, ∆narG/G2/G3; gray bar, ∆redD. Error bars indicate standard deviation (n = 3). (b) Phenotypes of hmpA deletion mutant. (c) Phenotypes of narGs mutants. Bottom, complementation of RED production and abnormal differentiation in the narG/G2/G3 mutant by exogenous NO2− (100 μM NO2− was added to the medium during culture). (d) Involvement of Nar in conversion of NO2− to NO. Cells were grown in the presence of 500 (left column) or 1,000 μM (right column) NO2−.
DevSR controls nitrogen oxide cycle and cellular NO levels
DevS is a heme-containing, NO-sensing histidine kinase17,18 that transduces signals to a transcriptional regulator DevR, constituting a two-component system (TCS) with DevS. Deletion of the S. coelicolor orthologs devS (SCO203) or devR (SCO204) (Supplementary Fig. 2) decreased transcripts of narG2 in S. coelicolor along with reducing Nar activity (Fig. 4a,b). Both the deletion mutants produced low levels of Red as observed in the ∆narG2 mutant and were recovered by a high concentration of NO2− (Fig. 4c). In addition, DevR (rDevR) protein could bind to the upstream region of narG2 in vitro (Fig. 4d). These results revealed that the S. coelicolor DevSR regulates the expression of narG2 and are integral components in the NO-forming pathway.
A DevSR two-component system controls narG2 gene expression in S. coelicolor.
Cells were grown on a cellophane membrane as in Fig. 1a for 72 h and transcriptional analysis of narG2 using real-time PCR (a) and measurement of total Nar activity in the membrane fraction (b) were performed. (c) devS and devR mutants exhibit the same phenotype as ∆narG2 (Fig. 3c) and the phenotype is canceled by exogenous 500 μM NO2−. (d) Reaction mixtures (10 μl) containing 400 nmol DIG-labeled oligonucleotide probe and recombinant DevR (0.1, 0.3, 0.5, 0.8 and 1.0 μg) were incubated for 30 min at 37 °C and separated by electrophoresis in a native 6% polyacrylamide gel. Unlabeled probe (200 ng) was used as competitor DNA and 200 ng of the control unlabeled oligonucleotide provided by DIG Gel Shift Kit 2nd generation was used as nonspecific DNA.
In vitro phosphorylation of rDevS, induced by its autokinase activity (Fig. 5a), was decreased in the presence of more than 1 μM NOC5. Exposure to high concentrations of exogenous NO2− or NOC5 also inhibited cellular transcripts of narG2 (Fig. 5b) and Nar activity (Supplementary Fig. 3). These results indicate that NO negatively regulates the DevSR TCS and transcription of the narG2 operon. The ∆hmpA mutant which accumulated more NO in the cell (Fig. 2a) excreted less NO2− into the medium after cultivation for 48 h (Fig. 5c) and the defect was complemented by introduction of hmpA gene (Fig. 5c). This observation supports the inhibitory effect of excess intracellular NO on narG2 expression since NO2− is a product of Nar reaction.
Endogenous NO controls its own intracellular concentration.
(a) Autophosphorylation of recombinant DevS on incubation with NOC5. Purified rDevS was incubated with 32P-γ-ATP in the presence of 0–10 μM NOC5 for the indicated time (10–30 s). (b) Effects of NO on the transcription of narG2 in the wild-type M145 strain. Cells were grown for 48 h as in Fig. 3a (left) and transferred to a fresh medium containing 100 μM NO2− or NO3− or the indicated concentration of NOC5 (0–10 μM); then cells were incubated for 1 h (NO2− or NO3−) or 25 min (NOC5) and total RNA was extracted from the cells and subjected to qPCR. Error bars indicate standard deviation (n = 3). (c) Excess endogenous NO stops NO2− production. Cells were grown and NO2− was stained as in Fig. 1c.
It can be concluded from these results that the DevSR TCS system regulates the concentration of endogenous NO by controlling the expression of the nar2 gene cluster and that NO itself acts as the negative regulator depending on its intracellular concentration.
NO2− is an intercellular signaling molecule
When the ∆narG/G2/G3 strain was cultured on a plate in which a single colony was surrounded by eight colonies of the parent strain M145, ∆narG/G2/G3 that had lost its Fhb-producing ability (Fig. 2) began to produce FhbA again (Fig. 6). This indicated that excreted NO2− or NO derived from NO2− acts as a signaling molecule for communication between cells and confirmed that NO is the end product of the nitrogen oxide cycle and is a hormone-like molecule. We found that Red-producing ability of the ∆narG/G2/G3 mutant strain was not restored (Supplementary Fig. 4), probably because NO2− transmitted to the mutant could not supply sufficient NO to trigger Red synthesis.
Cell-to-cell communication mediated by NO2−.
Spores of M145 or narGs mutant were inoculated onto the YEME-gln plate (a total of 9 spots) and incubated for 48 h. Only cells growing in the center spot were collected and subjected to western blotting to detect the production of Fhb. The M145/M145, M145 strain was inoculated at all 9 spots; ∆narGG2G3/M145; M145 was inoculated in all spots except the center spot in which the ∆narG/G2/G3 mutant was inoculated; the ∆narGG2G3/∆narGG2G3, ∆narG/G2/G3 mutant was inoculated at all spots.
Discussion
This study proposed a nitrogen oxide cycle that regulates cellular NO levels in S. coelicolor and its underlying metabolic and morphogenic mechanisms (Fig. 7). The homeostatic regulation of NO in cells is crucial to understanding the complex life of organisms. To date, endogenous production of NO without any exogenous nitrogen species is known to be achieved by transiently controlled production of NO synthase19. The S. coelicolor mechanism is unique in that Nar and Fhb play key roles in NO homeostasis. Conventional roles ascribed to Nar and Fhb are dissimilation of NO3− and detoxification of NO, respectively, both of which are mechanisms for responding to exogenously added nitrogen oxides in most bacteria. Recent studies showed a similar Nar-dependent NO2− production in human pathogen bacterium, Mycobacterium tuberculosis (Mtb). The NO2− production is believed to be a system for survival of Mtb in host20,21. We here demonstrated the role of Nar and Fhb in balancing the levels of cellular NO to control cell signaling in the proposed nitrogen oxide cycle mechanism (Fig. 7). The mechanism of S. coelicolor does not require exogenous nitrogen oxides, which highly suggests that it is not a mechanism for environmental responses, but is a constitutive housekeeping one.
Summary of NO metabolism in S. coelicolor A3(2) M145.
Nitrate reductase (Nar) and NO dioxygenase flavohemoglobin (Fhb) participated in NO2− and NO production, forming a nitrogen oxide cycle (NO3− → NO2− → NO → NO3−). NO controls its intracellular concentration by regulating Nar2 (narG2H2J2I2) and FhbA gene expressions via transcriptional factors NsrR and DevSR. Endogenously formed NO controlled antibiotic (RED) production and morphological differentiation.
In this study, we could not identify the mechanism involved in the production of NO3− in S. coelicolor. Our results indicated that NO2− and NO were generated after the production of NO3− (Fig. 2), indicating that NO3− is the first nitrogen oxide produced among the three nitrogen oxide species (NO3−, NO2− and NO) as the precursor of NO2− and NO. To date, few NO3− producing enzymes are known except for the enzymes that convert NO and NO2− to NO3−, thereby, indicating that NO3− generated in S. coelicolor was produced by some unidentified metabolic pathway. Therefore, we propose an extremely interesting topic regarding the production of NO3− in S. coelicolor.
While NOS and NO-generating nitrite reductase were previously known as the only enzymes to produce NO, recent studies have disclosed several other NO generation mechanisms. Nitric oxide generation from NO2− is known to depend on ether enzymatic or nonenzymatic reaction in bacterial cells. It was suggested that Escherichia coli and Salmonella typhimurium produce NO from NO2− by periplasmic cytochrome nitrite reductase22 and Nar23, respectively. Moreover, other heme-containing24,25,26 proteins (such as hemoglobins and NOS) and molybdenum proteins27,28 (such as aldehyde oxidase and xanthine oxidase) were shown to convert NO2− to NO. Further, nonenzymatic formation of NO from NO2− is also known29,30. Here we demonstrated the NOS-independent NO production in S. coelicolor (Fig. 2), its mechanism is to be elucidated.
Only links have been suggested between NsrR or DevSR and tolerance against stress8,31,32,33. In S. coelicolor, both proteins up- and down-regulate Fhb and Nar gene expressions respectively, to control cellular NO level in response to endogenously produced NO in an auto-regulation mechanism (Fig. 5). Despite NO production from early stages of culture, this mechanism can explain that NO performed as a signaling molecule at the later stage. Disruption of the genes devS or devR had a significant influence on secondary metabolism (Fig. 4c), indicating high stringency of this regulation system and the importance of endogenous NO. Generally, bacterial cells synchronously start new metabolic processes including secondary metabolism, which is called as quorum sensing34. Here, the excreted NO2− (or NO derived from NO2−) influenced FhbA production by neighboring cells (Fig. 6) showing that S. coelicolor cells communicate with one another via extracellular NO2− or NO derived from NO2−. Intracellular and extracellular NO2− are in equilibrium thus when extracellular NO2− concentrations increase, endogenous NO can overcome FhbA and the cell can start the new metabolic process which can shared between all cells. Thus, NO2− acts as an autoinducer in quorum sensing. So it should be NO2− that is also the purported signaling molecule.
Furthermore, we found a NO2− removal system (Fig. 5c). This NO homeostatic regulation system can explain the gradual decrease of accumulated NO2− (Fig. 1a). Thus, the NO2− removal system may be important not only to regulate NO homeostasis but also to complete the NO homeostatic regulation system in S. coelicolor. However, this notion needs to be proven by additional genetic studies.
The identification of NO as a signaling molecule in Streptomyces bacteria and the novel regulation system now allows us to take a step towards a better understanding of the regulation of synthesis of biologically active agents in the producer. In fact, our results show that the production of the antibiotic Red drastically increases depending on the concentration of exogenous NO (Fig. 3). The regulation of NO homeostasis in accordance with various systems continues to be an important subject for further investigation in all organisms to provide a new perspective on NO biology and to contribute towards human welfare.
Methods
Bacterial strains, plasmids and culture conditions
Strains used in this study are listed in Supplementary Table S1. Streptomyces coelicolor A3(2) M145 strain (wild-type) was obtained from the John Innes Centre, UK. Mannitol soya flour agar (2% mannitol, 2% soya flour, 2% agar) was used for sporulation and the strains were routinely grown on YEME-gln (glutamine) solid medium [0.3% yeast extract, 0.5% Bacto-peptone, 0.3% malt extract, 1% glucose, 50 mM L-glutamine (pH 7.2)] at 30 °C. The medium contained no detectable (by ion chromatography or colorimetric analysis) amount of nitrate or nitrite. Minimal solid medium (0.05% L-asparagine, 0.05% K2HPO4, 0.02% MgSO4· 7H2O, 0.001% FeSO4· 7H2O, 1% Glucose, 2% agar) was used for detection of NO2−_production ability in M145. E. coli DH5α (Takara, Kyoto, Japan) was used as the host for routine cloning. Media, culture conditions and DNA manipulations for Streptomyces and E. coli were performed as described by Kieser et al.35 and Green and Sambrook36, respectively. Media and culture conditions for the strains used for gene disruption followed a protocol of the REDIRECT PCR-targeting method37. E. coli HST04 dam-/dcm- (Takara) was used as a non-methylating cosmid and plasmid donor strain. E. coli Origami 2 (DE3) was used for recombinant DevS and DevR production. The plasmids, cosmids (kindly provided by the John Innes Centre, UK) and primers used for this study are listed in Supplementary Tables 2 and 3.
Construction of mutants and complementation
The open reading frames in the chromosomes were replaced with drug resistance cassettes by using REDIRECT PCR targeting37. Each drug resistance cassette flanked by Flippase recognition target (FRT) sites was amplified by PCR using PrimeSTAR GXL DNA polymerase (Takara) and each primer set is listed in Table S2. To obtain a target gene-disrupted version of the mutant cosmids by the λRed system, amplified cassettes were introduced into E. coli BW25113/pIJ79037 harboring an appropriate cosmid (Supplementary Table 2). The resulting construct was confirmed by PCR and introduced into E. coli HST04 dam-/dcm- (Takara) to obtain a non-methylating cosmid and each mutated cosmid was introduced into S. coelicolor A3(2) M145 or its derivatives by protoplast transformation. Drug‐resistant recombinants (Supplementary Table 1) were screened and successful recombination was checked by PCR using appropriate primer sets and a complementation study.
To obtain a marker-less mutant, the drug-resistance cassette was eliminated from the corresponding disrupted cosmid by introduction into E. coli strain BT340 in which recombination between both FRT mutagenesis cassette-flanking regions was induced by Flippase. In these new cosmids, only 81 base pairs (SCAR) remained in frame with the adjacent ORFs. Each resulting cosmid was introduced into the corresponding mutant and then the drug-sensitive mutant was screened and the replacement of drug-resistant cassette with SCAR was checked by PCR, using the appropriate primer set. Integration plasmids, pTYM19-narG2H2J2I2, pKU460-nsrR-hmpA and pKU460-devS-R, used for genetic complementation of knockout mutants, were prepared in the following manner. Each gene coding region, narG2H2J2I2 (SCO0216-0219), nsrR-hmpA (SCO7427-7428) and devS-R (SCO0203-0204), containing each upstream region (150–300 bp), was amplified by PCR, using the primer sets listed in Supplementary Table 3. The resulting narG2H2J2I2 fragment was cloned into the HinDIII site of pTYM1938. The nsrR-hmpA and devS-R fragments were cloned into the EcoRI/HinDIII site of pKU46039, respectively and the resulting plasmids were introduced into each disruptant.
Determination of NO3− and NO2−
NO3− was determined by ion chromatography, using a 761 Compact IC (Metrohm)6. NO2− was determined by Griess reagent assay40. Both nitrogen oxides were extracted from the medium for determination. Cells were grown for the indicated periods on a cellophane membrane covering the surface of a YEME-gln agar plate. After cultivation, cells on the cellophane were removed, five blocks of 1 × 1 cm were cut out from separate positions on the plate and the blocks were combined and homogenized in 5 ml distilled water. After centrifugation, the supernatant was further filtered through a 0.45-μm cellulose acetate filter and subjected to determination of nitrogen oxides in the culture medium. The determination depended on a standard curve made using medium containing 0, 1, 10, 25, 50 and 100 μM NO2− or NO3−.
Enzyme assay
Nar activity was assayed using dithionite/methylviologen as an electron donor, as previously described6.
Detection of FhbA by western blot analysis
Spores of M145 or its derivatives were inoculated on a solid medium plate at intervals of 1 cm (a total of 45 spots) with a toothpick. Culture conditions are presented in figure legends. Cells on the plate were incubated at 30 °C, harvested with a toothpick and disrupted by sonication in lysis solution (0.3-g urea, 0.15-g thiourea, 0.1-ml 20%CHAPS solution and 0.001-g DTT, in 1-ml distilled water). Seven micrograms of each soluble protein was separated by 12.5% SDS-PAGE, transferred onto an Immun-Blot PVDF Membrane (Bio-Rad) and probed with rabbit polyclonal FhbA antibody (1:2000). Proteins of interest were detected with goat anti-rabbit (GAR)-HRP conjugate (1: 1000, Bio-Rad) and visualized with the ECL Plus Western Blotting Detection System (GE), according to the provided protocol.
In situ detection of NO
Strains were grown at 30 °C on YEME-gln. Endogenously formed NO was detected using DAF-2DA (Dojindo) as described previously6. Photographs were taken with excitation at 495 nm and emission at 515 nm using FLUOVIEW FV300 System (Olympus).
Isolation of total RNA and qPCR
Total RNA was isolated using RNeasy Kit (Qiagen) from strains grown on cellophane-covered solid medium under several culture conditions, according to the manufacturer’s instructions. Conditions for each culture are given in the figure legends. cDNA was generated using a PrimeScript® RT reagent Kit with gDNA Eraser (Takara) and served as a template for qPCR. The primers used for qRT-PCR are indicated in Supplementary Table 3. qPCR was performed in a Thermal Cycler Dice Real Time System (Takara). PCR mixture (total 25 μl) contained 0.1 μg of generated cDNA, 10 pmol of an appropriate primer set (Table S3) and SYBR® Premix Ex TaqTMII (Takara). The hrdB gene of S. coelicolor was used as an internal control41.
Determination of RED
Undecylprodigiosin (RED) was determined as described42 with some modifications. Spores were inoculated onto a cellophane-covered YEME-gln plate at intervals of 1 cm (total of 45 spots) with a toothpick. After culture at 30 °C for 48 h, cells on cellophane were transferred to a fresh YEME-gln medium plate containing 0, 100, 500, or 1000 μM NO2−. Before the transfer, 300 μl distilled water was added onto the plate to allow tight contact of cellophane with the agar plate. Cells were incubated at 30 °C for 48 h and then collected and submitted for the determination of RED production. To remove the blue-pigmented antibiotic actinorhodin (ACT) from the cells, 1 M KOH was added and after incubation at 25 °C for 1 h, cells were centrifuged at 8,000 × g for 10 min and the supernatant containing ACT was removed. For RED, the cells were washed twice with 0.9% NaCl after ACT extraction. The resulting pellet was extracted with methanol (pH 2.0, adjusted with HCl) overnight at 25 °C, followed by centrifugation at 8,000 × g for 5 min and absorbance at 530 nm was measured. A molecular extinction coefficient of ε530 = 100,500 M−1cm−1 was used for the determination of RED.
Overexpression and purification of recombinant proteins
hmpA (SCO7428) encoding FhbA, devR (SCO7428) and devS (SCO7428) genes were amplified with primeSTAR GXL DNA polymerase, using primers listed in Supplementary Table 3. Each amplified gene was treated with NdeI and EcoRI and cloned into the corresponding sites of pET28b. Each expression construct was then transformed into E. coli Origami 2(DE3). For the overexpression of proteins, 1% of each overnight culture was inoculated into a fresh LB medium (1000 ml) containing 50 μM kanamycin and after 1 h growth at 37 °C, IPTG [0.2 mM (for FhbA and DevR) or 1 mM (for DevS)] was added to the medium. Each culture was then incubated at 30 °C (FhbA) or 16 °C (DevR and DevS) for 24 h. Grown cells were harvested and resuspended in Tris buffer [20 mM Tris-HCl (pH 8.0); containing 300 mM NaCl, 0.1 mM DTT, 20 mM imidazole and 10% glycerol] and broken by sonication on ice. After centrifugation at 4 °C, the supernatant was loaded onto a Ni-NTA Agarose column (Qiagen) and washed with the same buffer. Each target protein was eluted with Tris buffer containing 500 mM imidazole. Purified FhbA was used to produce rabbit polyclonal antibody.
Autophosphorylation of DevS
DevS (1 μg) was incubated with 10 μCi γ32P-ATP (Perkin-Elmer) in a 10-μl reaction mixture [containing 50 mM Tris HCl, 50 mM KCl, 5 mM MgCl2, and 10 μM ATP (Sigma, Missouri, USA) (pH 8.0)] at 30 °C for 10, 30 and 60 s, in the presence or absence of NOC5 (NO generator, half-life: 25 min) (Dojindo) with final concentrations of 0.1, 1 and 10 μM, respectively. Then, each mixture was promptly transferred to a heat-block set (56 °C) and 3 μl of stop solution (250 mM Tris-HCl, 10% SDS, 0.5% bromophenol blue, 50% glycerol, 500 mM DTT, pH 6.8) was added. After incubation for 10 min, each reaction mixture was separated by 12.5% SDS-PAGE and the gel was rinsed twice with water and subjected to autoradiography with BAS-2500 (Fuji film).
Other analytical methods
Cellular protein was determined using a protein assay reagent (Bio-Rad) after homogenization of cells with an ultra-sonicator as described previously6.
Additional Information
How to cite this article: Sasaki, Y. et al. Nitrogen oxide cycle regulates nitric oxide levels and bacterial cell signaling. Sci. Rep. 6, 22038; doi: 10.1038/srep22038 (2016).
References
Chandra, G. & Chater, K. F. Developmental biology of Streptomyces from the perspective of 100 actinobacterial genome sequences. FEMS Microbiol. Rev. 38, 345–379 (2014).
Crack, J. C. et al. Mechanistic insight into the nitrosylation of the [4Fe-4S] cluster of WhiB-like proteins. J Am Chem. Soc. 133, 1112–1121 (2011).
Saini, V., Farhana, A. & Steyn, A. J. Mycobacterium tuberculosis WhiB3: a novel iron-sulfur cluster protein that regulates redox homeostasis and virulence. Antioxid. Redox. Signal. 16, 687–697 (2012).
Singh, A. et al. Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc. Natl. Acad. Sci. USA 104, 11562–11567 (2007).
Smith, L. J. et al. Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive iron-sulfur cluster. Biochem. J. 432, 417–427 (2010).
Sasaki, Y., Takaya, N., Morita, A., Nakamura, A. & Shoun, H. Nitrite formation from organic nitrogen by Streptomyces antibioticus supporting bacterial cell growth and possible involvement of nitric oxide as an intermediate. Biosci. Biotechnol. Biochem. 78, 1603–1610 (2014).
Sasaki, Y., Takaya, N., Nakamura, A. & Shoun, H. Isolation of flavohemoglobin from the actinomycete Streptomyces antibioticus grown without external nitric oxide stress. Biosci. Biotechnol. Biochem. 68, 1106–1112 (2004).
Forrester, M. T. & Foster, M. W. Protection from nitrosative stress: a central role for microbial flavohemoglobin. Free. Radic. Biol. Med. 52, 1620–1633 (2012).
Zumft, W. G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616 (1997).
Kumon, Y. et al. Codenitrification and denitrification are dual metabolic pathways through which dinitrogen evolves from nitrate in Streptomyces antibioticus. J. Bacteriol. 184, 2963–2968 (2002).
Bentley, S. D. et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 (2002).
Fischer, M., Alderson, J., van Keulen, G., White, J. & Sawers, R. G. The obligate aerobe Streptomyces coelicolor A3(2) synthesizes three active respiratory nitrate reductases. Microbiology 156, 3166–79 (2010).
Fischer, M., Falke, D. & Sawers, R. G. A respiratory nitrate reductase active exclusively in resting spores of the obligate aerobe Streptomyces coelicolor A3(2). Mol Microbiol 89, 1259–73 (2013).
Tucker, N. P. et al. The transcriptional repressor protein NsrR senses nitric oxide directly via a [2Fe-2S] cluster. PLoS One 3, e3623 (2008).
Tucker, N. P., Le Brun, N. E., Dixon, R. & Hutchings, M. I. There’s NO stopping NsrR, a global regulator of the bacterial NO stress response. Trends Microbiol. 18, 149–156 (2010).
Takano, E. et al. Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2). Mol. Microbiol. 6, 2797–2804 (1992).
Lee, J. M. et al. O2- and NO-sensing mechanism through the DevSR two-component system in Mycobacterium smegmatis. J. Bacteriol. 190, 6795–6804 (2008).
Sardiwal, S. et al. A GAF domain in the hypoxia/NO-inducible Mycobacterium tuberculosis DosS protein binds haem. J. Mol. Biol. 353, 929–936 (2005).
Förstermann, U. & Sessa, W. C. Nitric oxide synthases: regulation and function. Eur. Heart. J. 33, 829–837, 837a–837d (2012).
Cunningham-Bussel, A., Zhang, T. & Nathan, C. F. Nitrite produced by Mycobacterium tuberculosis in human macrophages in physiologic oxygen impacts bacterial ATP consumption and gene expression. Proc Natl Acad Sci USA 110, E4256–65 (2013).
Cunningham-Bussel, A., Bange, F. C. & Nathan, C. F. Nitrite impacts the survival of Mycobacterium tuberculosis in response to isoniazid and hydrogen peroxide. Microbiologyopen 2, 901–11 (2013).
Corker, H. & Poole, R. K. Nitric oxide formation by Escherichia coli. J. Biol. Chem. 278, 31584–31592 (2003).
Gilberthorpe, N. J. & Poole, R. K. Nitric oxide homeostasis in Salmonella typhimurium: roles of respiratory nitrate reductase and flavohemoglobin. J Biol. Chem. 283, 11146–11154 (2008).
Cosby, K. et al. Nitrite reduction to nitric oxide by deoxyhemoglobin vasodilates the human circulation. Nat Med 9, 1498–505 (2003).
Shiva, S. et al. Deoxymyoglobin is a nitrite reductase that generates nitric oxide and regulates mitochondrial respiration. Circ Res 100, 654–61 (2007).
Vanin, A. F., Bevers, L. M., Slama-Schwok, A. & van Faassen, E. E. Nitric oxide synthase reduces nitrite to NO under anoxia. Cell Mol Life Sci 64, 96–103 (2007).
Li, H., Kundu, T. K. & Zweier, J. L. Characterization of the magnitude and mechanism of aldehyde oxidase-mediated nitric oxide production from nitrite. J Biol Chem 284, 33850–8 (2009).
Li, H., Samouilov, A., Liu, X. & Zweier, J. L. Characterization of the magnitude and kinetics of xanthine oxidase-catalyzed nitrate reduction: evaluation of its role in nitrite and nitric oxide generation in anoxic tissues. Biochemistry 42, 1150–9 (2003).
Lundberg, J. O., Weitzberg, E., Lundberg, J. M. & Alving, K. Intragastric nitric oxide production in humans: measurements in expelled air. Gut 35, 1543–6 (1994).
Zweier, J. L., Wang, P., Samouilov, A. & Kuppusamy, P. Enzyme-independent formation of nitric oxide in biological tissues. Nat Med 1, 804–9 (1995).
Ohno, H. et al. The effects of reactive nitrogen intermediates on gene expression in Mycobacterium tuberculosis. Cell Microbiol. 5, 637–648 (2003).
Park, H. D. et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48, 833–843 (2003).
Roberts, D. M., Liao, R. P., Wisedchaisri, G., Hol, W. G. & Sherman, D. R. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 279, 23082–23087 (2004).
Hong, K. W., Koh, C. L., Sam, C. K., Yin, W. F. & Chan, K. G. Quorum quenching revisited–from signal decays to signalling confusion. Sensors (Basel) 12, 4661–4696 (2012).
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces genetics (The John Innes Foundation, Norwich, 2000).
Green, M. & Sambrook, J. Molecular cloning: a laboratory manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York).
Gust, B., Challis, G. L., Fowler, K., Kieser, T. & Chater, K. F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100, 1541–1546.
Onaka, H., Taniguchi, S., Ikeda, H., Igarashi, Y. & Furumai, T. pTOYAMAcos, pTYM18 and pTYM19, actinomycete-Escherichia coli integrating vectors for heterologous gene expression. J. Antibiot. (Tokyo) 56, 950–956 (2003).
Komatsu, M., Uchiyama, T., Omura, S., Cane, D. E. & Ikeda, H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. USA 107, 2646–2651 (2010).
Griess, P. Bemerkungen zu der abhandlung der H. H. Weselsky und Benedikt “Ueber einige azoverbindungen.” Chem. Ber. 12, 426–428 (1879).
Aigle, B., Wietzorrek, A., Takano, E. & Bibb, M. J. A single amino acid substitution in region 1.2 of the principal sigma factor of Streptomyces coelicolor A3(2) results in pleiotropic loss of antibiotic production. Mol. Microbiol. 37, 995–1004 (2000).
Adamidis, T., Riggle, P. & Champness, W. Mutations in a new Streptomyces coelicolor locus which globally block antibiotic biosynthesis but not sporulation. J. Bacteriol. 172, 2962–2969 (1990).
Acknowledgements
This work was supported by a grant from the program for JSPS (Grant-in-Aid for Research Activity of Young Scientists B) to Y.S.
Author information
Authors and Affiliations
Contributions
Y.S. planned the studies. Y.S., T.N. and H.S. prepared the manuscript. Y.S., H.O., T.K., S.K., R.S., K.M., Y.Y., S.Y., S.I., K.O., K.O., H.I., H.T. and K.U. designed and performed the experiments. None of the authors have a financial interest related to this work.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Sasaki, Y., Oguchi, H., Kobayashi, T. et al. Nitrogen oxide cycle regulates nitric oxide levels and bacterial cell signaling. Sci Rep 6, 22038 (2016). https://doi.org/10.1038/srep22038
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep22038
This article is cited by
-
Mycothiol maintains the homeostasis and signalling of nitric oxide in Streptomyces coelicolor A3(2) M145
BMC Microbiology (2023)
-
Microbe-driven chemical ecology: past, present and future
The ISME Journal (2019)
-
Inter-Kingdom beach warfare: Microbial chemical communication activates natural chemical defences
The ISME Journal (2019)
-
Transcriptomic and fluxomic changes in Streptomyces lividans producing heterologous protein
Microbial Cell Factories (2018)
-
Characterization of a putative NsrR homologue in Streptomyces venezuelae reveals a new member of the Rrf2 superfamily
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.