Abstract
Differentiating neurons process the mechanical stimulus by exerting the protrusive forces through lamellipodia and filopodia. We used optical tweezers, video imaging and immunocytochemistry to analyze the role of non-muscle myosin-II on the protrusive force exerted by lamellipodia and filopodia from developing growth cones (GCs) of isolated Dorsal Root Ganglia (DRG) neurons. When the activity of myosin-II was inhibited by 30 μM Blebbistatin protrusion/retraction cycles of lamellipodia slowed down and during retraction lamellipodia could not lift up axially as in control condition. Inhibition of actin polymerization with 25 nM Cytochalasin-D and of microtubule polymerization with 500 nM Nocodazole slowed down the protrusion/retraction cycles, but only Cytochalasin-D decreased lamellipodia axial motion. The force exerted by lamellipodia treated with Blebbistatin decreased by 50%, but, surprisingly, the force exerted by filopodia increased by 20-50%. The concomitant disruption of microtubules caused by Nocodazole abolished the increase of the force exerted by filopodia treated with Blebbistatin. These results suggest that; i- Myosin-II controls the force exerted by lamellipodia and filopodia; ii- contractions of the actomyosin complex formed by filaments of actin and myosin have an active role in ruffle formation; iii- myosin-II is an essential component of the structural stability of GCs architecture.
Similar content being viewed by others
Introduction
During development, neurons are able to self-organize in precisely wired networks and are able to establish the appropriate synaptic connections. Neuronal navigation requires the existence of highly motile structures able to probe the mechanical properties of the surrounding environment and to search for the chemical cues leading to the formation of correct synaptic connections1,2. Neuronal exploration is guided by growth cones (GCs) located at the neurite tips3,4. GCs are composed of lamellipodia of different sizes, depending on the cell type and species from which thin filopodia with a submicron diameter emerge5. The primary source of motility in GCs is the polymerization of actin filaments6,7, controlled by a large set of regulatory proteins, such as Arp2/3, WASP, etc8 and molecular motors seem to participate in the overall process by controlling several aspects of the process.
The addition of actin monomers/oligomers to actin filaments in close contact with the membrane pushes the cellular membrane forward exerting a protrusive force6,9. An important determinant of force generation is the turnover of actin filaments, during which actin monomers or small oligomers are added to the barbed end of actin filaments (polymerization) and are removed from the other end (depolymerization). In this process the non-muscle myosin-II plays an important role: indeed myosin-II controls the retrograde flow of actin polymers by severing the actin filaments at their pointed end, providing the necessary treadmilling mechanism10. Myosins constitute a superfamily of motor proteins with major roles in several cellular processes such as cell adhesion, migration and division11. Myosin molecules, like all motor proteins, can walk along, propel and slide by other molecules and can produce tension on actin filaments. Generation of tension and force requires metabolic energy, usually provided by ATP hydrolysis and therefore myosins have appropriate catalytic sites in their amino-terminal (head) region. Myosin can associate to actin filaments to form the actomyosin complex, which can generate force. Like muscle myosin-II, non-muscle myosin-II (NMII) molecules are formed by three pairs of peptides with different molecular weight and function11. The three myosin-II isoforms NMIIA, NMIIB and NMIIC have similar structural and dynamical properties but have slightly different kinetics properties. Their major difference seems to reside in their regulation properties and different proteins control them through distinct phosphorylation sites11.
Myosin-II seems to be involved in the orchestration of actin polymerization/depolymerization but also of microtubules (MTs) dynamics. Indeed, it has been shown that actin oligomers driven by myosin-II interact with growing MTs and that myosin-II-dependent compressive force is necessary for MTs dynamics12 to form axons. The existence of a coupling between actin and MT dynamics is also supported by the observation that inhibition of myosin-II with Blebbistatin markedly accelerates axon growth and promotes the reorganization of both actin and MTs in GCs13. In this study we used Blebbistatin, selective potent inhibitor of myosin-II to assess the effect of myosin-II on the motility of the DRG GCs. Blebbistatin blocks the myosin in an ADP bound state which precedes the force generating step and therefore inhibits the actomyosin contraction14.
We have used Optical Tweezers (OT), to analyze the role of myosin-II in the force generation of DRG GCs lamellipodia and filopodia. OT provide a quantitative characterization of the exerted force with millisecond time resolution and pN sensitivity15. We have also used video imaging to characterize and quantify the 3D motion of lamellipodia, during which lamellipodia lift up vertically by some microns16. By combining these experimental methods with the use of inhibitors of cytoskeletal functions and of immunocytochemistry, we have explored the role of contractions of the actomyosin complex in the protrusion/retraction cycles, observed in lamellipodia of developing neurons. Here we confirm that myosin-II not only controls the retrograde flow of actin10 but it is also an essential component of the structural stability of GCs architecture regulating the coupling of actin filaments and microtubules dynamics and plays a fundamental role in the force generation of lamellipodia and - to some extent - also in filopodia.
Results
Large and highly motile lamellipodia emerge from dissociated neurons from DRG after 6–12 hours of culture17,18,19. These lamellipodia can exert forces larger than 20 pN and their leading edge can move with a speed of 30–100 nm/s19. In our preparation, motility is restricted to lamellipodia and filopodia of dissociated neurons from DRG, which do not migrate and their soma remains approximately in the same position on the dish for several hours. After 2–3 days of culture, dissociated neurons establish physical contacts and motility of lamellipodia and filopodia is reduced. Therefore, we analyzed the effect of inhibitors of, myosin-II (Blebbistatin), actin polymerization (Cytochalasin-D) and microtubule polymerization (Nocodazole) on lamellipodia and filopodia after 24–48 hours of culture, when their motility is more pronounced.
The effect of Blebbistatin, Cytochalasin-D and Nocodazole on lamellipodia protrusion/retraction cycles
Lamellipodia emerging from the soma of DRG neurons protrude and collapse continuously and their protrusion/retraction cycles were followed by video imaging (see Methods). By analyzing these image sequences with Algorithm I, described in the Methods section, the average distance of the lamellipodium leading edge was measured from a reference point (C) chosen at the base of the lamellipodium (see Methods) and the periods of protrusion/retraction cycles were calculated (red bars in Fig. 1). When 30 μM Blebbistatin was added to the medium bathing of the neuronal culture, protrusion/retraction cycles of lamellipodia could be observed but with a period 30–50% longer than in control condition and after 15 minutes lamellipodia shrank (Fig. 1a and 1b). When a higher concentration of Blebbistatin was used, such as 100 μM, lamellipodia shrank within 2–3 minutes and motility was completely suppressed.
The effect of Blebbistatin, Cytochalasin-D and Nocodazole on protrusion/retraction cycles.
(a) Cycles of protrusion/retraction of lamellipodia vs time. The dotted line represents the time of inhibitor addition. (b) Average periods of lamellipodia protrusion/retraction cycles in control condition (red) and in the presence of 30 μM Blebbistatin (Bleb 30 μM, blue), from n = 8 neurons. Student t-test showed that the data significantly differs with respect to control, P < 0.005. Data represents mean ± SEM. (c) The fraction of pixels in focus of lamellipodia in different focal planes (h = 2, 3 and 4 μm) above the coverslip. Data averaged from 8 experiments. The vertical bar indicates the SEM and the vertical broken line indicates the time at which the drug was added. (d–e) Images of lamellipodia emerging from a DRG neuron in control condition (d) and after treatment with 25 nM Cytochalasin-D (CD 25 nM) (e), Scale bar, 5 μm. (f–h) As in (a–c) but in the presence of 25 nM Cytochalasin-D, from n = 10 neurons. (h) as in (c) but for 10 experiments. (i–j) Images of lamellipodia emerging from a DRG neuron in control condition (i) and after treatment with 500 nM Nocodazole (Noco 500 nM,). (k–m) As in (a–c) but in the presence of 500 nM Nocodazole, from n = 8 neurons. (m) As in (c) but for 7 experiments. All the data were checked with chi-square test for Normal distribution before applying the student's t test.
In control condition, during protrusion/retraction cycles, lamellipodia also moved upwards by 2–5 μm: indeed, at a focal plane 3 or 4 μm above the coverslip their leading edge could be seen well in focus. By using Algorithm II described in the Methods section, the number of pixels of a lamellipodium in focus at different heights, i.e. at 2, 3 and 4 μm above the coverslip, was counted and followed in time (Fig. 1c). This algorithm allowed quantifying the extent of the axial motion and the effect of different inhibitors of cytoskeletal proteins on this axial motion. After the addition of 30 μM Blebbistatin to the bathing medium, the period of protrusion/retraction cycles increased from an average of 96.1 ± 3.3 s in control condition to 136.7 ± 5.9 s (Fig. 1b). Lamellipodia not only prolonged the duration of their protrusion/retraction cycles (Fig. 1a and b) but also reduced the average height reached during these cycles in the presence of 30 μM Blebbistatin (Fig. 1c). Indeed, the fraction of pixels in focus at 2 μm above the coverslip increased, while those in focus at 3 and 4 μm above the coverslip decreased (Fig. 1c).
Treatment with a concentration of 100 μM Blebbistatin invariably led to the suppression of lamellipodia motility. We also investigated the effect of other inhibitors known to affect and abolish motility, but acting on different biochemical targets. Cytochalasin-D is a well-known and specific inhibitor of actin filament polymerization20. Cytochalasin-D bound to the barbed end of actin filaments blocking the addition of new actin monomers or oligomers. Concentrations of Cytochalasin-D, such as 50 or 100 nM caused lamellipodia to shrink completely and abolished almost entirely the GCs motility, confirming the fundamental role of actin filament polymerization. Nocodazole inhibits the microtubules polymerization21 and, in our experiments, lamellipodia motion was almost entirely abolished in the presence of 1 μM of Nocodazole. Blebbistatin, Cytochalasin-D and Nocodazole reduced lamellipodia motility but did not have the same effect on lamellipodia and filopodia morphology: lamellipodia treated with Cytochalasin-D shrank and showed the formation of small ruffles but did not acquire the ‘filopodish’ appearance (Fig. 1d and e) observed in lamellipodia treated with Blebbistatin (see Discussion). Lamellipodia treated with Nocodazole shrank showing neither small ruffles nor the filopodish appearance as seen in Cytochalasin-D and Blebbistatin respectively (Fig. 1i and j). Addition of 25 nM Cytochalasin-D and 500 nM Nocodazole did not abolish the protrusion/retraction cycles (Fig. 1f and k).
The period of protrusion/retraction cycles increased from an average of 95.2 ± 7.3 s in control condition to 131.5 ± 9.8 s in the presence of 25 nM Cytochalasin-D (Fig. 1g) and 110.9 ± 5.0 s in the presence of 500 nM Nocodazole (Fig. 1l). The same concentration of Cytochalasin-D also reduced the ability of treated lamellipodia to lift up along the vertical direction during these protrusion/retraction cycles: the fraction of edges seen in focus at focal planes higher than 3 μm significantly decreased and lamellipodia edges seen in focus at a plane 2 μm above the coverslip became much more frequent (Fig. 1h). The application of 500 nM Nocodazole caused a transient shrinkage of GCs, but, within a couple of minutes, lamellipodia were able to lift up in the vertical direction almost as in control condition (Fig. 1m). This differential effect of Blebbistatin and Cytochalasin-D versus Nocodazole indicates a major role of myosin-II and actin polymerization in lamellipodia axial motion and a minor role of microtubules.
The effect of Blebbistatin, Cytochalasin-D and Nocodazole on the force exerted by lamellipodia
Having analyzed the effect of Blebbistatin, Cytochalasin-D and Nocodazole on the kinetics of protrusion/retraction cycles, we used OT to analyze changes of the force exerted by lamellipodia and filopodia caused by these inhibitors. Untreated lamellipodia pushed trapped beads (Fig. 2a–c) exerting maximum forces up to 10–20 pN as previously described22 and often a bead could be displaced out of the optical trap. Lamellipodia of DRG treated with 30 μM Blebbistatin could also pull and push a trapped bead (Fig. 2e–g) but with a lower force (Fig. 2h).
The effect of Blebbistatin, Cytochalasin-D and Nocodazole on the force generated by lamellipodia.
(a) Low-resolution image of a bead trapped in front of a lamellipodium emerging from the soma of a DRG neuron in control condition. Scale bar, 5 μm (b–c) High-resolution images during a push. At t1 the bead is in the optical trap (b) and when the lamellipodium grows, at t2, it pushes the bead (c). The cross indicates the center of the optical trap. Scale bar, 1 μm. (d) The three components Fx, Fy and Fz of the force exerted when the lamellipodium pushes the bead. (e–h) As in (a–d) but in the presence of Blebbistatin (Bleb 30 μM). (i–l) As in (a–d) but in the presence of Cytochalasin-D (CD 25 nM). (m–p) As in (a–d) but in the presence of Nocodazole (Noco 500 nM). The trap stiffness is kx,y = 0.10 pN/nm., kz = 0.03 pN/nm. (q–t) Comparison of the force exerted by lamellipodia in control condition (red), 30 μM Blebbistatin (blue), 25 nM Cytochalasin-D (black) and 500 nM Nocodazole (green) and in all four different stereotyped behaviors: LP (lateral push), LR(lateral retraction), VP (vertical push) and VR (vertical retraction). In each case, by using the student t-test, the force measured in the presence of inhibitors was lower than the one measured in control condition with a significance *P < 0.005. Data represent mean ± SEM. (u–x) Average Fv relationship, (FV)avg, normalized to Fmax for VP(u), LP(v), VR (w) and LR (x). All the data were checked with chi-square test for Normal distribution before applying the student's t test.
The addition of 25 nM Cytochalasin-D and 500 nM of Nocodazole caused lamellipodia to shrink, reduced neuronal motility and the amplitude of generated forces (Fig. 2i–p). In several experiments we were able to measure the maximum force exerted by the same lamellipodia in control condition and in the presence of inhibitors. These measurements were then divided into four different stereotyped behaviors: vertical push (VP), vertical retraction (VR), lateral push (LP) and lateral retraction (LR), where vertical refers to the push or pull of the bead in the axial direction (perpendicular to the plane of the coverslip) and lateral refers to the push or pull of the bead in the lateral direction (parallel to the plane of the coverslip) (Fig. 2q–t). In these experiments all tested inhibitors reduced the force by about 50%, in case of LP. While in case of LR, Blebbistatin and Cytochalasin-D reduced the force by 40–50%, but Nocodazole reduced the force by 75% compared to control condition. For VP and VR the force was decreased in Blebbistatin and Cytochalasin-D by about 50% and in Nocodazole by more than 80% compared to control condition (Table 1). In the great majority of experiments, treatment with 25 nM Cytochalasin-D and 500 nM of Nocodazole for longer than 30–50 minutes completely abolished GCs motility.
The average Fv relationships (FV)avg were computed from the measured displacements (see Methods). Fv relationships obtained from a single experiment were normalized to Fmax and averaged to obtain average Fv relationships, (FV)avg19. At the beginning, the bead was in the trap far from the lamellipodia and its velocity was zero. During push the lamellipodia leading edge moved toward the trapped bead with constant velocity. Before coming to a solid contact with the bead, the bead velocity increased but later - after complete contact – beads and lamellipodia moved with the same velocity. Therefore (FV)avg relationships after an initial rise of v exhibited a flat shape, during which the mean velocity remained constant while the force increased (Fig. 2u–x). The analysis of the Force-velocity (Fv) relationships (Fig. 2u–x) shows that both inhibitors did not modify the shape of the Fv relationships but reduced the maximal velocity v for both vertical and lateral pushes and retractions. Lamellipodia velocity was reduced more potently by 500 nM Nocodazole than 25 nM Cytochalasin-D and 30 μM Blebbistatin (compare green, black and blue traces in Fig. 2 u–x).
These results show that Nocodazole, Blebbistatin and Cytochalasin-D reduce the maximal force exerted by protruding lamellipodia and the maximal velocity of their leading edges.
Changes of noise during force generation with Blebbistatin and Cytochalasin-D
A remarkable feature of force generation during vertical and lateral push is the concomitant increase of noise when the lamellipodia push the bead18. This increase of noise is not present when the lamellipodium retracts, pulling the bead away from the optical trap. We have previously shown18 that in controlled GCs, the relation between the variance of the measured displacement σ2 and the exerted force F is upward convex and σ2 increases from about 50 nm2 to 150 nm2 as the force also increases (Fig. 3a, b and c, red traces) and that this increase of σ2 is abolished by Jasplakinolide, inhibiting actin filament depolymerisation23. In GCs treated with 12.5 and 25 nM Cytochalasin-D the relation between F and σ2 was flat and almost no increase of σ2 was observed even when the force exceeded 8 pN (grey and black traces in Fig. 3a). In the presence of 30 μM Blebbistatin, a small increase of σ2 from about 40 to 60 nm2 was observed (blue trace in Fig. 3b). In case of 500 nM Nocodazole σ2 increased from 30 to 70 nm2 (green trace in Fig. 3c).
The effect of Blebbistatin and Cytochalasin-D on the elementary events underlying force generation.
(a) Average force – variance relationship for lateral pushes in control condition (red curve) and in the presence of Cytochalasin-D (CD 25 nM, black and grey curves). (b) As in (a) but in the presence of Blebbistatin (Bleb 30 μM, blue curve). (c) As in (a) but in the presence of Nocodazole (Noco 500 nM, green curve) (d–f) Magnification of the z component during push in the presence of Cytochalasin-D (d), in the presence of Blebbistatin (e) and in the presence of Nocodazole (f). Original traces were filtered by the nonlinear diffusion algorithm, resulting in a smooth component and jumps. Jumps were not detected frequently during a push in the presence of Cytochalasin-D and Nocodazole but more often during a push in the presence of Blebbistatin. (g–i) Density of forward j+ and backward j− jumps during pushes in the presence of Cytochalasin-D (g), in the presence of Blebbistatin (h) and in the presence of Nocodazole (i). Because of a residue noise, jumps with an amplitude lower than 2 nm could not be detected.
Following bead adhesion to the lamellipodium membrane18, σ2 could decrease to less than 6 nm2 and subsequently, when the lamellipodium pushed the bead, forward and backward jumps constituting the elementary events underlying force generation appeared. In the presence of 500 nM Nocodazole, 25 nM Cytochalasin-D, forward and backward jumps could be observed but were less frequent than in control condition (Fig. 3d and f). Also in the presence of 30 μM Blebbistatin forward and backward jumps were observed and were more frequent (Fig. 3e and h) than those observed in the presence of Cytochalasin-D. The amplitude of forward j+ and backward jumps j− were exponentially distributed (Fig. 3g, h and i) and were fitted by the equations A+ e−j+/j+* and A− e−j-/j−* where A+ and A− are the frequency of forward and backward jumps, respectively and j+* and j−* are the mean amplitude of forward and backward jumps, respectively. Mean values of these parameters obtained in control condition and in the presence of Nocodazole, Cytochalasin-D and Blebbistatin are shown in Table 2. In control condition the mean values of j+* and j−* were 5.1 ± 1.3 and 4.9 ± 1.2 nm respectively with corresponding rates A+ and A− of 157.3 ± 12.2 and 155.5 ± 11.1 events/s respectively. In the presence of both Blebbistatin and Cytochalasin-D the mean values of forward and backward jumps j+* and j−* decreased by about 50%, in agreement with the reduced or absence of noise which increased during force generation caused by the addition of the two inhibitors (Fig. 3a and b). These inhibitors, however, had a different action on the jump frequency: larger concentrations of Cytochalasin-D progressively reduced A+, i.e. the rate of the appearance of forward jumps but not of backward jumps, in agreement with the known effect of Cytochalasin-D that blocks actin filament polymerization20. Blebbistatin reduced both the forward and backward rates A+ and A−. In the presence of Nocodazole the mean values of the forward and backward jumps j+* and j−* remained the same, but the jump frequency was reduced by more than 60% (Fig. 3c and Table 2).
Blebbistatin makes filopodia able to exert a larger force
Nocodazole, Blebbistatin and Cytochalasin-D reduced the amplitude of the force exerted by DRG lamellipodia, but, rather surprisingly, we observed that the force exerted by filopodia treated with Blebbistatin was larger than in untreated filopodia.
In control condition, when filopodia emerged from lamellipodia (Fig. 4a), they moved randomly in space searching for chemical cues before they retracted. These filopodia could exert forces very rarely exceeding 4 pN when a trapped bead was kept in their random motion (Fig. 4b). From the same neurons, the force exerted by filopodia after the addition of 30 μM Blebbistatin was measured (Fig. 4c–d). In these conditions, filopodia emerging from lamellipodia that had shrunk were still able to exert a force which was often larger (Fig. 4d) and were also able to exert a significant force along a vertical direction (compare red traces in Fig. 4b and d). Collected data from 12 neurons show that the average force exerted by filopodia was 2.7 pN in control condition and increased to 4.2 pN in the presence of 30 μM Blebbistatin. However, filopodia in the presence of 500 nM Nocodazole together with 30 μM Blebbistatin exerted forces of 2.6 ± 0.2 pN (Fig. 4h) similar to those observed in control condition.
The effect of Blebbistatin on the force exerted by DRG filopodia.
(a) Images of a bead trapped in front of a filopodium emerging from a GCs of DRG neuron. At t1 the bead is in the optical trap and at t2–t3 the filopodium pushes the bead. The cross indicates the centre of the optical trap. (b) The three components Fx, Fy and Fz of the force exerted by the filopodium. (c–d) As in (a–b) but in the presence of Blebbistatin (Bleb 30 μM, blue). (e–f) As in (a–b) but in the presence of Nocodazole (Noco 500 nM). (g–h) As in (a–b) but in the presence of Nocodazole+Blebbistatin (Blebb+Noco). (i) Filopodia force in Control, in presence of Blebbistatin, Nocodazole and Nocodazole+Blebbistatin. By using the student t-test, the force measured in the presence of inhibitors was lower than the one measured in control condition with a significance *P < 0.005. Data represent mean ± SEM. The trap stiffness is kx,y = 0.10 pN/nm, kz = 0.03 pN/nm. All the data were checked with chi-square test for Normal distribution before applying the student's t test.
These results show that Blebbistatin reduces the amplitude of the force exerted by lamellipodia but increases the force exerted by filopodia of DRG neurons (Fig. 4i); this increase of the force exerted by filopodia is abolished by the concomitant application of Nocodazole.
Discussion
The present manuscript describes the effect of the inhibition of myosin-II on the morphology, kinetics and dynamics of lamellipodia and filopodia emerging from the soma and GCs of DRG neurons. Our results confirm that myosin-II not only controls the retrograde flow of actin10 but also controls and regulates the structural stability of GCs architecture managing the coupling of actin filaments and microtubules dynamics. Our results also show that the contractions of the actomyosin complex formed by filaments of actin and myosin have an active role during lamellipodia retractions. Let us now discuss more in detail these issues.
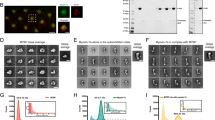
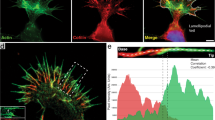
There are three isoforms of myosin-II in GCs, which have often a different localization in GCs13,24 possibly underlying different functions25,26. We examined the localization of NMIIA and NMIIB in DRG GCs by immunostaining. We determined simultaneously the cellular distribution of actin, tubulin and one of the two myosin isoforms, i.e. NMIIA and NMIIB (see Fig. SI1). The staining for NMIIB was preferentially localized in the central domain and transition zone of the GCs, in agreement with previous observations10,13 and very rarely we detected staining in the filopodia. In contrast, we observed a more diffuse staining of NMIIA, present in the central and transition zone of the GCs, but also in its periphery, near its leading edge and occasionally also in some filopodia. We analyzed also the actin and tubulin distribution in lamellipodia emerging from the soma of differentiating DRG neurons. Lamellipodia sprouting from the soma had an extensive network of actin filaments interspersed with rare filaments of microtubules. Also in these lamellipodia staining of NMIIA was clearly present at their leading edge, while staining for NMIIB was more restricted near the soma and rarely extended to the leading edge of lamellipodia.
After treatment with 20–50 μM Blebbistatin, a powerful inhibitor of both myosin-II isoforms27, lamellipodia emerging from the soma and from GCs distant from the soma, changed their morphology, lost their sheet-like structure and appeared ‘filopodish’ (Fig. 5a–d). After Blebbistatin treatment (Fig. 5d–f) sparse actin filaments were clearly visible and they did not appear to be joined by the usual actin network. Untreated GCs at the tip of long neurites had the core of microtubules surrounded by a mesh of actin filaments and very rarely microtubules entered the filopodia, which were primarily composed of actin filaments. After treatment with Blebbistatin, the terminal ends of neurites were not only composed of actin filaments but also of microtubules at the most distant GCs tips (Fig. 5e and f). The average number of filopodia per GCs in untreated DRG neurons was 7.5 ± 1.2 and was 6.8 ± 1.2 after treatment with 30 μM Blebbistatin (Fig. 5g).
The effect of Blebbistatin on GCs morphology.
(a–b) Lamellipodium emerging from a DRG neuron in control condition and after treatment with 30 μM Blebbistatin, respectively. Note the ‘filopodish’ appearance of the lamellipodia after Blebbistatin treatment. (c) Immunostaining of DRG lamellipodium in control condition for actin (green) and tubulin (blue) staining. (d) As in (c) but in the presence of 30 μM Blebbistatin. (e) Immunostaining of a GC after Blebbistatin treatment for actin, NMIIA and tubulin and merge of the three staining. Arrows and arrowheads indicate filopodia with and without a clear staining for tubulin, respectively. (f) Immunostaining of a GC after Blebbistatin treatment for actin, NMIIB and tubulin and merge of the three staining. (g) The average number of filopodia per GC before (red) and after treatment with Blebbistatin (blue). (h) The fraction of filopodia with a staining for microtubules in control condition (red bar) and after Blebbistatin treatment (blue bar). Student t-test showed that data significantly differ when compared to the control, P < 0.05. All data were checked with chi-square test for Normal distribution before applying the student's t test. Scale bar, 5 μm.
If the mean number of filopodia per GC was not significantly affected by myosin-II inhibition, treatment with Blebbistatin had a profound effect on the distribution of microtubules inside the filopodia: in control condition the fraction of filopodia emerging from GCs exhibiting a staining for microtubules was 0.07 ± 0.02 (Fig. 5h, red bar) but after Blebbistatin treatment it increased to 0.42 ± 0.04 (Fig. 5h, blue bar), showing that inhibition of NMII elevated the presence of microtubules inside filopodia. Filaments of NMII could cross-link actin filaments providing the network with a diffuse lateral connectivity gluing together the sparse actin filaments resulting in a sheet-like overall structure. Inhibition of NMII destroyed this connectivity leading to the observed ‘filopodish’ appearance.
Contractions of the actomyosin complex play a fundamental role in several cellular processes such as changes of the cellular shape28, cell migration25,29 cell-cell and cell-matrix adhesion30, cell division and cell differentiation11. During the cycles of protrusion/retraction here analyzed (Fig. 1) lamellipodia ruffle after their maximal protrusion. These events seem to precede the usual lamellipodium retraction and, given the localization of NMIIA at the lamellipodium periphery, they are most likely originated from contractions of the actomyosin. These observations suggest a dual and complementary role for the two myosin-II isoforms: NMIIA located also at the periphery of lamellipodia, undergoing ruffle formation, could mediate a contraction of the actomyosin complex initiating retraction and NMIIB located more centrally near the transition region of the lamellipodium could control actin turnover10. Numerical simulations of the actomyosin complex have shown that generated stresses are overwhelmingly contractile and force chains play a major role31,32,33.
The ruffle formed during the retraction of the lamellipodia could be the artifact of the 2D substrate. Lamellipodia ruffle forms because of inefficient formation of focal adhesion34, while in 3D matrices the motility of the cell switches between adhesion-dependent mesenchymal (elongated) and adhesion-independent amoeboid (rounded) cell motility35.
When NMII was inhibited by Blebbistatin, we observed two significant morphological changes: lamellipodia lost their sheet-like appearance and became ‘filopodish’ (Fig. 5a and b) and filopodia emerging from GCs had a higher proportion of microtubules inside (Fig. 5g and h) in agreement with previous findings36. These morphological changes were mirrored by the observation that filopodia treated with Blebbistatin exert a larger force (Fig. 4). The mean flexural rigidity of microtubules is 2.2 × 10−23 Nm2 which is almost 1000 times larger than that of actin filaments and equals to 7.3 × 10−26 Nm237: therefore, filopodia from GCs treated with Blebbistatin are expected to have a larger stiffness and to exert a larger force. When microtubule polymerization was concomitantly inhibited by Nocodazole (Fig. 4), filopodia exerted a force comparable to that observed in control condition.
These observations are consistent with the emerging view that inhibition of NMII promotes axon regeneration13. Chondroitin sulfate proteoglycans (CSPGs), major components of the extracellular matrix in the CNS, inhibit axonal regeneration after injury, through the activation of NMII by phosphorylation of regulatory myosin light chain (RLC) ultimately remodeling cytoskeletal dynamics38. Inhibition of NMII by Blebbistatin promotes axon outgrowth irrespective of the presence of CSPGs in both CNS and PNS neurons13,38 providing therefore a promising pharmacological/chemical treatment for neuronal regeneration.
The results reported in the present manuscript confirm the essential role of NMII in cytoskeletal dynamics and in the orchestration of both actin and MT dynamics in GCs12,13,38. As shown in Fig. 5, after Blebbistatin treatment, the proportion of filopodia with MTs inside them increases from 0.07 to 0.42 suggesting that Blebbistatin has facilitated the growth of MTs filaments. The biochemical pathway through which NMII affects MT dynamics is not known and it is probably not involving the Rho-kinase (ROCK)13: indeed, inhibition of NMII promotes axon growth but not the inhibition of the Rho-ROCK pathway. On the other hand, repulsive guidance molecule (RGMa) induces neurite outgrowth inhibition through RhoA and Rho-kinase dependent phosphorylation of NMIIA RLC resulting in F-actin reduction39. These findings suggest, therefore, mechanistically distinct actin- and MT-based GC responses.
Methods
Neuron preparation
Wistar rats at postnatal days 10 to 12 (P10–P12) were sacrificed by decapitation after anesthesia with CO2 in accordance with the Italian Animal Welfare Act. The Ethics Committee of the International School for Advanced Studies (SISSA-ISAS) has approved the protocol (Prot.n. 2189-II/7). After dissection, Dorsal Root Ganglias (DRG) were incubated with trypsin (0.5 mg/ml; Sigma-Aldrich, Milan, Italy), collagenase (1 mg/ml; Sigma-Aldrich) and DNase (0.1 mg/ml; Sigma-Aldrich) in 5 ml Neurobasal medium (Gibco, Invitrogen, Milan, Italy) in a shaking bath (37°C, 35–40 min). After mechanical dissociation, they were centrifuged at 300 rpm, resuspended in culture medium and plated on poly-L-lysine-coated (0.5 μg/ml; Sigma-Aldrich) coverslips. Neurons were incubated for 24 h to 48 h and nerve growth factor (50 ng/ml; Alomone Labs, Jerusalem, Israel) was added before performing the measurements.
Quantification of lamellipodia motility
Z-stack phase contrast imaging was performed to quantify the kinetics of protrusion/retraction cycles of lamellipodia in lateral and axial directions. Stacks of images at a frequency of 0.1–1 Hz were acquired. Every stack of images was composed of an image, focused at the plane containing the coverslip where neurons were cultured and images focused at 1,2,3,4,5 and 6 microns above the coverslip. Two algorithms were developed: Algorithm I was designed to quantify in a semi-automatic way the time course of protrusion/retraction cycles and Algorithm II was designed to quantify the vertical motion of lamellipodia during these cycles.
Algorithm I
Images focused on the coverslip plane at different times of the protrusion/retraction cycles (Fig. 6a, t1–t3) were analyzed: edges were extracted using standard procedures40 and the contour of the neuron was obtained (red line in Fig. 6b). A reference point at the base of the lamellipodium was selected (red cross in Fig. 6b) and an angle covering the lamellipodium was drawn (green shadow in Fig. 6b). The mean distance between the red cross and the points forming the detected contour inside the green shadow was computed and plotted (Fig. 6c). In this plot, representing the mean distance of the lamellipodium leading edge from the reference point, local maxima and minima were detected (green and red asterisks, respectively, in Fig. 6c). The interval between a successive green and red point was taken as the period of that protrusion/retraction cycle.
Characterization of lamellipodial protrusion/retraction cycles and of vertical motion.
(a) From left to right: three images of the lamellipodium undergoing cyclic waves of protrusion (t2) and retraction (t1 and t3) in control condition; the white dotted line represents the leading edge of the lamellipodia. Scale bar, 5 μm. (b) Diagram of the method used for the semi-automatic detection of protrusion/retraction cycles. See text for more details. (c) Time evolution of the distance of the lamellipodium leading edge from the reference point indicated by the red cross in (b). Local maxima and minima represent maximal protrusion and retraction, respectively. (d) Stack of 6 images acquired from 6 focal planes at distance h from the coverslip where neurons were cultured. Scale bar, 5 μm. Red and blue arrows indicate section of lamellipodia above and below the focused plane, respectively. The pixels above focus appear brighter and the pixels below appear darker. (e) Fractional pixels in focus of lamellipodia in different focal planes (h = 2, 3 and 4 μm) above the coverslip indicating the fractional reached height by lamellipodia. The continuous solid lines are smoothing over a time window of 100 s.
Algorithm II
Algorithm II was based on classical depth-from-focus algorithms introduced in Computer Vision16 to recover 3D information from stacks of images acquired at different focal planes. These algorithms were used to restore the lamellipodia motion in the vertical direction. Briefly, for each pixel (i,j) and for each image I(i,j,h) acquired at a focal plane h microns above the coverslip, the gradient ∇ I(i,j,h) was computed. The point at location (x,y) has the height h if the feature at point (x,y) is in focus on the plane h, determined as the plane for which ∇ I(i,j,h) has the maximum value. Images of the neuron taken from different focal planes separated by 1 μm are shown in Fig. 6d (h = 1,…,6 μm) from which ∇I(i,j,h) was computed.
In order to characterize the vertical motion ability of a lamellipodium for each value of h, we computed the fraction of pixels - in a given region of interest - in focus at the height h (Fig. 6e). In this way we could quantify the effect of Blebbistatin, Cytochalasin-D and Nocodazole on the ability of lamellipodia to lift up vertically.
Force Measurements
The Optical Tweezers (OT) set-up used for force measurements, the procedures followed to compute the Force-Velocity (Fv) relations and the elementary events were as previously described18,19. The optical tweezers set-up was built as described in Ref. 22.
Computation of Fv relationships
The velocity v = (vx, vy, vz) of the bead was obtained by numerical differentiation of its sampled position x = (x(n), y(n), z(n)) n = 1,…N. Numerical differentiation was computed either by convolution of the position components x(n), y(n) and z(n) with the derivative of a Gaussian filter 1/[σ(2π)1/2] exp(−t2/σ2) (Gaussian filtering) or by Linear regression. Gaussian filters corresponding to cut-off frequencies of 0.2, 1 and 10 Hz were used. Further details can be found in Ref. 19.
Jumps determination by non linear diffusion filtering
In order to detect jumps, we used an algorithm based on non linear diffusion41,42. The algorithm is based on the Toolbox of Frederico D'Almeida (see http://www.mathworks.com/matlabcentral/fileexchange/3710-nonlinear-diffusiontoolbox). Further details can be found in Ref. 18.
Immunostaining
Cells were fixed in 4% paraformaldehyde containing 0.15% picric acid in phosphate-buffered saline (PBS), saturated with 0.1 M glycine, permeabilized with 0.1% Triton X-100, saturated with 0.5% BSA in PBS (all from Sigma-Aldrich, St.Louis, MO) and then incubated for 1 h with primary antibodies: mouse monoclonal antibody against neuronal class III β-tubulin-TUJ1 (Covance, Berkeley, CA) and rabbit polyclonal antibodies against myosin-IIA and IIB (both from Sigma-Aldrich, St.Louis, MO). The secondary antibodies were goat anti-rabbit 594 Alexa (Invitrogen, Life Technologies, Gaithersburg, MD, USA) and anti-mouse IgG2a biotynilated (Santa Cruz Biotechnology, Santa Cruz, CA) and the incubation time was 30 min. F-actin was marked with Alexa Fluor 488 phalloidin, whereas biotin was identified by Marina Blue-Streptavidin (Invitrogen, Life Technologies, Gaithersburg, MD, USA) and incubated for 30 min. All the incubations were performed at room temperature (20–22°C). Cells were examined using a Leica DMIRE2 confocal microscope (Leica Microsystems GmbH, Germany) equipped with DIC and fluorescence optics, diode laser 405 nm, Ar/ArKr 488 nm and He/Ne 543/594 nm lasers. The fluorescence images (1024 × 1024 pixels) were collected with a 63× magnification and 1.3 NA oil-immersion objective. Leica LCS Lite and Image J by W. Rasband (developed at the U.S. National Institutes of Health and available at http://rsbweb.nih.gov/ij/) were used for image processing.
References
Ghashghaei, H. T., Lai, C. & Anton, E. S. Neuronal migration in the adult brain: are we there yet? Nat. Rev. Neurosci. 8, 141–51 (2007).
Solecki, D. J., Govek, E.-E. & Hatten, M. E. mPar6 alpha controls neuronal migration. J. Neurosci. 26, 10624–5 (2006).
Song, H. & Poo, M. The cell biology of neuronal navigation. Nat. Cell Biol. 3, E81–8 (2001).
Goodman, C. Mechanisms and molecules that control growth cone guidance. Annu. Rev. Neurosci. 19, 341–377 (1996).
Mongiu, A. K., Weitzke, E. L., Chaga, O. Y. & Borisy, G. G. Kinetic-structural analysis of neuronal growth cone veil motility. J. Cell Sci. 120, 1113–25 (2007).
Mogilner, a. & Oster, G. Cell motility driven by actin polymerization. Biophys. J. 71, 3030–45 (1996).
Pollard, T. D. & Borisy, G. G. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–65 (2003).
Pak, C. W., Flynn, K. C. & Bamburg, J. R. Actin-binding proteins take the reins in growth cones. Nat. Rev. Neurosci. 9, 136–47 (2008).
Raucher, D. & Sheetz, M. P. Cell spreading and lamellipodial extension rate is regulated by membrane tension. J. Cell Biol. 148, 127–36 (2000).
Medeiros, N. A., Burnette, D. T. & Forscher, P. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat. Cell Biol. 8, 215–26 (2006).
Vicente-Manzanares, M., Ma, X., Adelstein, R. S. & Horwitz, A. R. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10, 778–90 (2009).
Burnette, D. T. et al. Myosin II activity facilitates microtubule bundling in the neuronal growth cone neck. Dev. Cell 15, 163–9 (2008).
Hur, E.-M. et al. Engineering neuronal growth cones to promote axon regeneration over inhibitory molecules. Proc. Natl. Acad. Sci. U. S. A. 108, 5057–62 (2011).
Allingham, J. S., Smith, R. & Rayment, I. The structural basis of blebbistatin inhibition and specificity for myosin II. Nat. Struct. Mol. Biol. 12, 378–9 (2005).
Bustamante, C., Macosko, J. C. & Wuite, G. J. Grabbing the cat by the tail: manipulating molecules one by one. Nat. Rev. Mol. Cell Biol. 1, 130–6 (2000).
Krotkov, E. Focusing. Int. J. Comput. Vis. 1, 223–237 (1988).
Amin, L., Ercolini, E., Shahapure, R., Migliorini, E. & Torre, V. The role of membrane stiffness and actin turnover on the force exerted by DRG lamellipodia. Biophys. J. 102, 2451–60 (2012).
Amin, L., Ercolini, E., Shahapure, R., Bisson, G. & Torre, V. The elementary events underlying force generation in neuronal lamellipodia. Sci. Rep. 1, 153 (2011).
Shahapure, R., Difato, F., Laio, A. & Bisson, G. Force generation in lamellipodia is a probabilistic process with fast growth and retraction events. Biophys. J. 98, 979–88 (2010).
Cooper, J. A. Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 105, 1473–8 (1987).
Dent, E. & Kalil, K. Axon branching requires interactions between dynamic microtubules and actin filaments. J. Neurosci. 21, 9757–9769 (2001).
Cojoc, D. et al. Properties of the force exerted by filopodia and lamellipodia and the involvement of cytoskeletal components. PLoS One 2, e1072 (2007).
Bubb, M. R. Effects of Jasplakinolide on the Kinetics of Actin Polymerization. An explanation for certain in vivo observations. J. Biol. Chem. 275, 5163–5170 (2000).
Wylie, S. R. & Chantler, P. D. Myosin IIC: a third molecular motor driving neuronal dynamics. Mol. Biol. Cell 19, 3956–68 (2008).
Betapudi, V. Myosin II motor proteins with different functions determine the fate of lamellipodia extension during cell spreading. PLoS One 5, e8560 (2010).
Wylie, S. R. & Chantler, P. D. Separate but linked functions of conventional myosins modulate adhesion and neurite outgrowth. Nat. Cell Biol. 3, 88–92 (2001).
Kovács, M., Tóth, J., Hetényi, C., Málnási-Csizmadia, A. & Sellers, J. R. Mechanism of blebbistatin inhibition of myosin II. J. Biol. Chem. 279, 35557–63 (2004).
Roh-Johnson, M. et al. Triggering a cell shape change by exploiting preexisting actomyosin contractions. Science 335, 1232–5 (2012).
Solecki, D., Trivedi, N. & Govek, E. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron 63, 63–80 (2009).
Pasapera, A. M., Schneider, I. C., Rericha, E., Schlaepfer, D. D. & Waterman, C. M. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol. 188, 877–90 (2010).
Kim, T. Determinants of contractile forces generated in disorganized actomyosin bundles. Biomech. Model. Mechanobiol. (2014) 10.1007/s10237-014-0608-2.
Lenz, M., Gardel, M. L. & Dinner, A. R. Requirements for contractility in disordered cytoskeletal bundles. New J. Phys. 14 (2012).
Dasanayake, N. L., Michalski, P. J. & Carlsson, A. E. General Mechanism of Actomyosin Contractility. Phys. Rev. Lett. 107, 118101 (2011).
Borm, B., Requardt, R. P., Herzog, V. & Kirfel, G. Membrane ruffles in cell migration: indicators of inefficient lamellipodia adhesion and compartments of actin filament reorganization. Exp. Cell Res. 302, 83–95 (2005).
Ulrich, T. A., Jain, A., Tanner, K., MacKay, J. L. & Kumar, S. Probing cellular mechanobiology in three-dimensional culture with collagen-agarose matrices. Biomaterials 31, 1875–84 (2010).
Rösner, H., Möller, W., Wassermann, T., Mihatsch, J. & Blum, M. Attenuation of actinomyosinII contractile activity in growth cones accelerates filopodia-guided and microtubule-based neurite elongation. Brain Res. 1176, 1–10 (2007).
Gittes, F., Mickey, B., Nettleton, J. & Howard, J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J. Cell Biol. 120, 923–34 (1993).
Yu, P., Santiago, L. Y., Katagiri, Y. & Geller, H. M. Myosin II activity regulates neurite outgrowth and guidance in response to chondroitin sulfate proteoglycans. J. Neurochem. 120, 1117–28 (2012).
Kubo, T. et al. Myosin IIA is required for neurite outgrowth inhibition produced by repulsive guidance molecule. J. Neurochem. 105, 113–26 (2008).
Marthon, Ph., Thiesse, B. & Bruel, A. Edge Detection By Differences Of Gaussians. in Proc. SPIE 0595 318–327 (1986). 10.1117/12.952277.
Perona, P. & Malik, J. Scale-Space and Edge-Detection Using Anisotropic Diffusion. Ieee Transactions on Pattern Analysis and Machine Intelligence. 12, 629–639 (1990).
Weickert, J. Applications of nonlinear diffusion in image processing and computer vision. ACTA MATHEMATICA UNIVERSITATIS COMENIANAE 70, 33–50 (2001).
Acknowledgements
We acknowledge the financial support of the following projects within the Seventh Framework Programme for Research of the European Commission: the FOCUS Project n. FP7-ICT-270483, the NEUROSCAFFOLDS Project n. 604263. We thank M. Lough for editing the paper.
Author information
Authors and Affiliations
Contributions
W.S. and L.A. equally contributed to this work. V.T. conceived the project and designed the experiments. W.S., L.A. and E.E. performed the experiments and W.S., L.A. analyzed the data. W.S., L.A., E.E. and P.F. contributed to reagents/materials/analysis tools. V.T., W.S. and L.A. wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Sayyad, W., Amin, L., Fabris, P. et al. The role of myosin-II in force generation of DRG filopodia and lamellipodia. Sci Rep 5, 7842 (2015). https://doi.org/10.1038/srep07842
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07842
This article is cited by
-
The growth determinants and transport properties of tunneling nanotube networks between B lymphocytes
Cellular and Molecular Life Sciences (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.