Abstract
The geographical distributions of diploid and polyploid Galax urceolata overlap in the Blue Ridge Mountains, USA. As part of an investigation into the evolutionary forces governing the establishment of polyploids and their coexistence with diploids, we examined the population frequencies of diploids, triploids and tetraploids in the area of overlap. Ploidy was inferred from estimates of DNA content, using flow cytometry, for 1570 individuals sampled from 42 populations. Across the entire sampling area, diploids and tetraploids were most abundant (55% and 34% of individuals, respectively), whereas triploids were least abundant (11%). Cytotype frequencies differed significantly among the northern, central and southern regions of the range (G = 649.02, d.f. = 4, P < 0.0001), with diploids most frequent in the north-east and least frequent in the south-west. Twenty-six per cent of the populations contained three cytotypes, 33% contained two, and 40% had a single cytotype. Populations with two cytotypes occurred in all possible cytotype combinations, but when triploids were present, they were always in the minority. Uniform populations were either diploid (81%) or tetraploid (19%), but never triploid. Overall, populations are predominantly diploid or tetraploid but rarely evenly mixed, suggesting disruptive selection for chromosome number in G. urceolata. The contribution of ecological sorting and frequency-dependent mating success to the distribution of polyploids and diploids is discussed.
Similar content being viewed by others
Introduction
Polyploidy, or chromosome multiplication, is a central feature of many plants. Estimates vary, but between 47 and 70% of extant angiosperm species are believed to be polyploid (Masterson, 1994). Because of its broad effects on a variety of plant attributes, polyploidy is an important mechanism of range expansion and local adaptation (Levin, 1983), and through its influence on reproductive isolation is arguably one of the most important mechanisms of plant species diversification (Levin, 1983). Despite its taxonomic and evolutionary significance, little is known of the processes governing the origins and establishment of polyploids in natural populations (Thompson & Lumaret, 1992; Ramsey & Schemske, 1998).
Geographical distributions of polyploids and their diploid progenitors provide useful insights into the population biology of polyploidy. Differences in geographical range among cytotypes may reflect historical patterns of colonization or genetic differentiation in ecological amplitude that has occurred in association, or subsequent to, polyploid formation (Levin, 1983). Where cytotype ranges overlap, local distributions offer opportunities to examine the interactions between cytotypes and to infer the evolutionary forces influencing the establishment of polyploids and coexistence with diploids in natural populations (Lewis & Suda, 1976). However, relatively few studies have described the frequencies of diploid and polyploid cytotypes and the pattern of differentiation within geographical areas of overlap (Soltis, 1984; Van Dijk et al., 1992). Studies of this nature are particularly important, because describing population cytotype structure is a crucial first step in understanding the possible evolutionary forces governing the establishment of cytotypes.
Evolutionary forces governing the frequency and distribution of polyploids in regions of mixed cytotype can be divided into two kinds: (i) ecological sorting in heterogeneous environments (Endler, 1977; Fowler & Levin, 1984; Van Dijk & Bijlsma, 1994); and (ii) frequency-dependent mating success (Levin, 1975; Felber, 1991; Arnold, 1997). These two models represent forces acting on cytotype distribution, through viability and fertility selection, respectively, and are not mutually exclusive. Although their underlying mechanisms and evolutionary implications may differ, both models make similar predictions as to what pattern of polyploid variation should be observed in natural populations. Specifically, populations should be highly differentiated with respect to ploidy, containing predominantly diploid or tetraploid individuals. This prediction is testable empirically, and has generated interest in examining the distribution of cytotypes in natural populations (Van Dijk & Bijlsma, 1994; Felber-Girard et al., 1996).
Galax urceolata (Poiret) Brummitt is a perennial plant, endemic to the Appalachian mountain range in south-eastern United States (Gleason, 1952). The only member of its genus, G. urceolata, is described as an autopolyploid and exhibits variation in chromosome number throughout much of its geographical range (Baldwin, 1941; Stebbins, 1971; Nesom, 1983). Individuals are either diploid (2n = 2x = 12), triploid (2n = 3x = 18) or tetraploid (2n = 4x = 24). However, there is little information on the frequency of cytotypes within and among populations. As part of a larger investigation into the evolution of polyploidy in this species, our objectives here were to describe the frequencies of cytotypes within populations and the differentiation among populations in the geographical region of overlap. Specifically, we asked the following questions. (i) What are the frequencies of diploid, triploid and tetraploid plants? (ii) Do populations consist of single or multiple cytotypes? (iii) Do cytotypes in mixed populations coexist at nearly equal frequencies or are populations dominated by a single cytotype?
One factor contributing to the paucity of large-scale surveys of chromosomal variation is the practical difficulty involved in counting chromosomes for large numbers of individuals (Michaelson et al., 1991). A technique that may permit such large-scale studies is flow cytometry, an automated procedure that estimates DNA content per nucleus. Using this method, it is possible to evaluate the DNA content of thousands of nuclei per individual in a few minutes (Arugumanathan & Earle, 1991; Costich et al., 1993) and thereby infer ploidy. In this study, we used this technique to describe the cytotype distribution of G. urceolata. Because its application to population biology has only recently been explored (Keeler, 1989; Costich et al., 1993), we had two additional objectives: (i) to quantify the relationship between DNA content, measured using flow cytometry, and ploidy, based on chromosome counts; and (ii) to test for geographical variation in DNA content among populations of the same ploidy.
Materials and methods
Study organism
Galax urceolata is a perennial herb found in moist or dry mixed deciduous forests from West Virginia to Alabama, USA (Gleason, 1952). The plant has long-petioled basal, evergreen leaves and is thought to reproduce both asexually via stolons, and sexually via erect scapes bearing a long spike-like raceme of small white flowers. To date there is no published research on the breeding system of G. urceolata, but floral morphology, pollen/ovule ratios and pollinator observations suggest that it may be partially outcrossing (T. L. Burton, unpubl. data).
Sampling
Cytotype frequencies were estimated for 42 populations of G. urceolata located along the Blue Ridge Mountains in Virginia, North Carolina, Tennessee and Georgia, USA (Fig. 1). Earlier surveys of chromosome number by Nesom (1983) and Baldwin (1941) showed that the Blue Ridge Mountains represent the centre of the geographical range for G. urceolata and encompass the region of overlap for diploid and tetraploid cytotypes. In this study, populations were located and sampled at 20 km intervals along the Blue Ridge Parkway and connecting roads. Young leaves were sampled throughout each population. Because G. urceolata can grow clonally, leaves were sampled every 1.5–2 m to maximize the number of genets encountered. Samples were taken throughout the entire population and therefore sample sizes were proportional to the areal extent of the population. The number of ramets sampled per population ranged from three to 150, for a total range-wide sample of 1570. Leaves were stored in plastic bags on ice in the field and then refrigerated until DNA content could be measured.
Map of the geographical distribution of Galax urceolata showing the population frequencies of diploid, triploid and tetraploid individuals across the entire range. Pie diagrams represent the proportion of diploid (black), tetraploid (white) and triploid (striped) cytotypes in each population. Numbers indicate the sampling locations of G. urceolata for this study.
Estimation of DNA content
To determine the ploidy of each plant, DNA content was estimated using flow cytometry. Leaves (≈2–3 cm2) were chopped with a razor blade on ice in the presence of cold buffer containing 15 mM HEPES, 1 mM EDTA, 80 mM KCl, 20 mM NaCl, 300 mM sucrose, 0.2% Triton X-100, 0.1% β-mercaptoethanol and 0.5 mM spermine (Bino et al., 1992), and then subsequently filtered through a 30 μm nylon mesh. The samples were then centrifuged at high speed (2000g) for 10 s and resuspended in 320 μL of ice-cold buffer consisting of 10 mM MgSO4.7H2O, 1 mg/mL dithiothreitol, and 10% w/v Triton X-100, 2% propidium iodide stock solution (5 mg/mL) (Solution ‘A’, Arugumanathan & Earle, 1991) and 0.24% DNAase-free RNAase (Boehringer Mannheim, cat. no. 1119915).
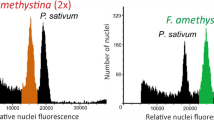
The samples of resuspended nuclei were analysed using an EPICS Profile flow cytometer (Coulter Electronics, Haileah, Florida) equipped with an Argon ion laser (25mW, Omnichrome 150) emitting at 488 nm. For each sample, ≈10 000 events were recorded and displayed as a histogram. Chicken red blood cells (CRBC) were used as an external standard for DNA content determination and run after every 5–10 samples. To account for any variation caused by daily differences in flow cytometer calibrations, fluorescence per sample was converted to a relative fluorescence value by expressing the sample as a proportion of the mean fluorescence of the CRBC standard analysed during the same run (hereafter referred to as relative fluorescence).
Relationship between DNA content and ploidy
To use DNA content as a measure of ploidy, we first examined the relationship between DNA content and ploidy (based on chromosome counts). In 1995, chromosome counts were made on 141 individuals, representing all three cytotypes, from 14 populations. Leaf samples for flow cytometry were collected at the same time as tissue for chromosome counts. Leaves were collected from the same individual, or from leaves less than 25 cm apart, to ensure they represented the same genet. Chromosomes were counted in young leaves using the methods of Warmke (1935) and Nesom (1983). Young leaves were sampled from each plant, fixed in 3:1 ethanol:acetic acid and kept at 4°C for 72 h. Upon return from the field, leaf samples were transferred to 70% ethanol. Each leaf was placed in acetocarmine stain and brought to a boil five times. Tissue was gently chopped with a new razor blade and squashed on a glass slide in acetic acid. Two to five cells at metaphase were observed under oil immersion (100× mag.) and chromosomes counted. An analysis of variance (ANOVA), was used to examine the differences in DNA content among ploidy levels.
Assigning ploidy
The relative fluorescence values for 141 known diploids, triploids and tetraploids sampled in 1995 served as standards for classifying the remaining samples. With these data, specific relative fluorescence ranges for all three cytotypes were identified. The ranges of relative fluorescence values were 1.8–3.1 for diploids and 3.6–6.2 for tetraploids. The relative fluorescence values of triploids fell between diploids and tetraploids in all but five cases. In these five cases, the relative fluorescence value of known triploids overlapped with the range of relative fluorescence values of known diploids and tetraploids (relative fluorescence values for these triploids were 2.6, 3.6, 3.6, 4.0 and 4.2, respectively). For this reason, the range of values for triploids was set to include any individual with a relative fluorescence value greater than the maximum value for known diploid individuals (3.1) and less than the minimum value for known tetraploid individuals (3.6). Truncating the range in this way provides a minimum estimate of the frequency of triploids.
Because estimates of DNA content using flow cytometry can vary between runs and therefore between years (Costich et al., 1993; Husband & Schemske, 1998), it was necessary to confirm that these expected ranges were applicable to samples analysed in 1996/97. To do this, we examined the range of relative fluorescence values for populations in a region that had been described as uniformly diploid (populations 23, 24, 37 and 38) (Nesom, 1983). The range of relative fluorescence values for these pooled populations was 1.9–2.9. In addition, we measured the relative fluorescence of individuals from population 8, a population determined by us in 1995 to be uniformly diploid. The relative fluorescence range was 1.8–3.1, indicating that our estimates of the range of relative fluorescence were consistent among years.
The analysis of 141 individuals for chromosome number and fluorescence indicated that genome size varied within a ploidy level. To determine whether DNA content of diploids and tetraploids differs among populations, we estimated the relative fluorescence values for each ploidy in all populations containing at least three individuals of the same ploidy. Mean relative fluorescence values among populations within each ploidy level were compared using a Kruskal–Wallis test.
Geographical variation in cytotype composition
The number and frequency of cytotypes per population were estimated for the whole range (n = 42 populations), and for three regions representing northern, central and southern portions of the range (Fig. 1). The region boundaries were set perpendicular to the Blue Ridge Parkway transect and were positioned to give an equal number of populations in each region. The northern region was located north of the border between Virginia and North Carolina, and included populations 18, 19, 22, 23, 24, 25, 35, 36, 37, 38, 39 and 40 (n = 12). The central region occurred north of a line that runs parallel to the North Carolina–South Carolina border, and bisects populations 7 and 8. The central region included populations 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 21, 33, 34 and 42 (n = 14). The southern region occurred south of the line between populations 7 and 8 and included populations 1, 3, 4, 5, 6, 7, 20, 28, 29, 30, 31, 32 and 44 (n = 13). Populations 26, 27 and 41 were excluded from some of the following analyses (where indicated) because these populations were located off the Blue Ridge Mountain range and were not in the mixed cytotype zone according to Nesom (1983) and Baldwin (1941). The number of cytotypes per population and the frequencies of diploid, triploid and tetraploid individuals for each region were compared using G-tests. Variation among regions in the mean frequency of diploid and tetraploid individuals for each population in each region was examined using an analysis of variance (ANOVA).
To test the predictions that populations should be strongly differentiated for cytotype, populations were divided into five categories according to the percentage of diploid or tetraploid individuals (0–20%, 20–40%, 40–60%, 60–80% and 80–100%) and analysed using a chi-squared goodness-of-fit test to determine if there was deviation from a uniform distribution. The same analysis was repeated using the percentages of tetraploids, as these frequencies are independent of diploids.
Results
Relative fluorescence and chromosome number
The relationship between DNA content (based on relative fluorescence) and chromosome number was estimated using 44 diploid, 13 triploid and 84 tetraploid individuals, originally assigned ploidy levels using chromosome counts. Mean relative fluorescence was 2.55 (SE ± 0.34) for diploids, 3.62 (SE ± 0.44) for triploids and 5.57 (SE ± 0.70) for tetraploids. Means differed significantly from each other in an ANOVA and Scheffés multiple comparison test (F2,198 = 391.6, P < 0.0001). The tetraploid/diploid ratio for relative fluorescence was 2.18. The observed mean relative fluorescence for triploids (mean = 3.62) was less than the expected value (4.06) based on the mean of the diploid and tetraploid values. Relative fluorescence values for diploids corresponded to a mean 2C DNA content of 5.78 pg.
The population mean relative fluorescence for diploids ranged from 2.19 (SE ± 0.04) to 2.78 (SE ± 0.30) and differed significantly among the five populations examined (Kruskal–Wallis test H = 10.98, P < 0.05) (Table 1). Mean fluorescence values for tetraploid populations ranged from 4.63 (SE ± 1.05) to 5.89 (SE ± 0.06) and also differed significantly among populations (H = 12.82, P < 0.05). In diploids and tetraploids, mean population fluorescence was highest in the most southern population on the sampling transect and increased monotypically towards the northern part of the range.
Cytotype structure
Diploid, triploid and tetraploid cytotypes were all observed within the study area. In the entire sample of 1570 individuals, 55% were diploid, 34% were tetraploid and 11% were triploid (Table 2). There were significant differences in the frequencies of cytotypes among the three regions (G = 649.02, df = 4, P < 0.0001), with the frequency of diploids being highest in the northern region (0.89) and lowest in the southern region (0.36). In contrast, the frequencies of triploids and tetraploids were highest in the southern region (0.15 and 0.49, respectively) and lowest in the northern region (0.08 and 0.02, respectively).
Within the geographical range of G. urceolata sampled, populations consisted of three, two or one cytotype (Fig. 1). Of the 42 populations, 26% contained three cytotypes and 33% contained two (Table 3). Forty per cent of all populations contained a single cytotype. Of the 14 populations consisting of two cytotypes, 46% contained diploids and triploids, 23% were composed of triploids and tetraploids and 31% contained diploids and tetraploids. Among populations that consisted of a single cytotype, 81% were diploid and 19% were tetraploid (Table 3). No uniform triploid populations were found.
The proportion of populations containing three, two or one cytotype differed significantly among the three regions (G = 9.85, d.f. = 4, P < 0.05) (Table 3). Populations with three cytotypes were least common in the northern region (0.08) and most common in the southern region (0.30). In contrast, the proportion of populations with two cytotypes was highest in the central region (0.57) and lowest in the northern and southern regions (0.25 and 0.23, respectively). Of the three kinds of populations comprising two cytotypes, only the diploid–triploid cytotype combination was seen in the north, whereas all three types of population were found in the central and southern regions. In addition, the frequency of uniform populations that were diploid declined from one in the northern region to 0.50 in the southern region.
The mean coefficient of variation (CV) in the frequency of diploids among populations within regions was 0.81, almost two times the CV for variation among regions (0.49). The CV among populations differed among regions, with the northern region exhibiting greater homogeneity (CV = 0.23) than the central and southern regions (CV = 1.08 and 1.13, respectively).
The distribution of cytotype frequencies within populations was not uniform. On average, populations that consisted of three cytotypes contained more triploids and tetraploids than diploids (Table 4). In populations that consisted of two cytotypes, whenever triploids were present they were in the minority. When populations consisted of only diploid and tetraploid individuals, the diploids were in the majority. The frequency of triploids in a population differed depending on the other constituents of the population. Triploids were more likely to occur in populations with both diploid and tetraploid cytotypes than in populations with either diploids or tetraploids alone (G = 6.33, d.f. = 1, P < 0.05).
In an analysis of all 42 populations throughout the range of G. urceolata, diploids tended to be either very common or very rare, but not intermediate in frequency (χ2 = 8.4, d.f. = 4, P < 0.01). The analysis of the distribution of tetraploids indicated that they also are very common or very rare, but rarely intermediate (χ2 = 8.2, d.f. = 4, P < 0.01) (Fig. 2). Collectively these data showed that populations of G. urceolata are usually predominantly diploid or tetraploid with few evenly mixed populations.
Discussion
Galax urceolata is a valuable system for studying polyploid evolution in natural populations for a number of reasons. First, based on cytological, morphological and biochemical criteria, it has been described as an autopolyploid (Stebbins, 1950; Soltis et al., 1983; Epes & Soltis, 1985). This conclusion and the fact that Galax is a monotypic genus, suggests that diploid G. urceolata is the sole progenitor of the tetraploid cytotype. Our evidence that the DNA content of tetraploids is approximately twice the DNA content of the diploids is consistent with this idea, although it is unclear how frequently and by what pathways polyploids have arisen (Ramsey & Schemske, 1998). Secondly, the distributions of diploid and tetraploid G. urceolata overlap for most of their geographical ranges (Nesom, 1983), providing a unique opportunity to examine the evolutionary forces governing the origin and establishment of polyploids and the conditions favouring their coexistence. As a first step towards this goal, we examined the geographical distribution of diploids and polyploids, and quantified the variation within and between populations, throughout the geographical range of overlap. Below, we examine the results in the light of recent theory, and consider the implications for the origin of polyploids and the maintenance of mixed cytotype regions. We begin by evaluating the use of flow cytometry for overcoming some of the practical limitations of studying chromosomal polymorphism on a large geographical scale.
In this study, we estimated frequencies of polyploidy in populations of G. urceolata using flow cytometry. Using this method, ploidy for 1570 individuals was inferred from measures of DNA content. Such an approach proved extremely effective, given that several thousand nuclei could be examined in each individual and that there was a strong association between DNA content and ploidy. Distinguishing triploids from diploid and tetraploid individuals was sometimes problematic. This ambiguity is significant given that observations of triploids are conflicting in previous studies (Baldwin, 1941; Nesom, 1983). To minimize upward bias of triploid frequencies, we chose to truncate the range of triploid values and thereby estimate the number of triploid individuals conservatively. Although this is not the first application of flow cytometry to a population-level survey (Keeler, 1989; Husband & Schemske, 1998), the number of cases is limited and to our knowledge surveys using flow cytometry have never been carried out on such a large scale.
Although measures of DNA content using flow cytometry were sufficient for identifying chromosome cytotypes, we also found that a significant portion of the variation in DNA content could not be accounted for by differences in ploidy. Rather, DNA content estimates varied widely among populations within a given ploidy. Population DNA content differed significantly among five populations containing diploids and among five populations containing tetraploids within the sample area. The underlying determinants of this variation are unknown, but may be associated with genetic instabilities influencing the number (e.g. aneuploidy) and size (e.g. DNA fragment loss or gain; Song et al., 1995) of chromosomes (Price, 1988), or measurement error caused by variation in machine running conditions and the degree of intercalation of fluorescent dye. Because we used a DNA standard for each run and ensured that concentrations of propidium iodide were at saturating levels, we feel the latter possibility is unlikely. The association between DNA content and geographical position observed for both diploids and tetraploids suggests that either changes in DNA are environmentally induced (Evans, 1968) or selection may be favouring decreased DNA content at higher latitudes, within a given ploidy. Interestingly, the relationship between DNA content and latitude is opposite to other studies conducted on a larger spatial scale (Price, 1988). In any event, variation in DNA content among populations of the same ploidy did not confuse the classification of individuals, as this variation was considered in the identification of the ranges of relative fluorescence values for diploid, triploid and tetraploid individuals.
Population differentiation for cytotype
Cytotype frequencies were not consistent throughout the geographical range of overlap in G. urceolata. Diploids, for example, decreased from north to south, whereas the frequency of triploids and tetraploids increased from south to north throughout the range. In addition, uniform diploid populations were the only type of single cytotype population in the northern region; however, they declined to 67% in the central region and 50% in the southern region, where uniform diploid and tetraploid populations occurred in equal proportions. The gradual change in mean cytotype frequency from the northern to the southern region represents a weak clinal pattern throughout the range. However, within each region, populations differed widely in cytotype number and frequency. In fact, the coefficient of variation for the frequency of diploids among populations within regions was almost twice as great as the CV among regions, and there was no evidence of any gradual change in cytotype composition with geographical position. Therefore, the distribution of cytotypes at the population level is more accurately described as a mosaic, with considerable spatial variation in cytotype composition (Harrison & Rand, 1989). This mosaic structure has also been reported in other studies of geographical variation and population structure of polyploid taxa (Husband & Schemske, 1998).
Within the range of overlap, populations of G. urceolata were strongly differentiated for chromosome number, with either diploids or tetraploids being predominant. This result is consistent with theoretical models that predict that populations at equilibrium should consist primarily of diploids or tetraploids (Levin, 1975; Fowler & Levin, 1984; Felber, 1991). This prediction is founded on the premise that mating is random and fitness in mixed cytotype populations is frequency-dependent. Cytotypes in the minority will be at a mating and transmission disadvantage, because they are less likely to encounter mates of the same cytotype.
A similar trend towards monotypic populations might be expected to occur if fitness is determined primarily by habitat, and cytotypes are ecologically differentiated from one another. In this case, populations will be nearly uniform and the distribution of diploid and tetraploid populations should reflect the geographical pattern of environmental heterogeneity. The information available on habitat differentiation in diploid and tetraploid G. urceolata is conflicting, and insufficient to assess the role of ecological factors affecting fitness and cytotype distribution. No quantitative evidence for ecological differentiation has been collected; however, Nesom (1983) indicated that, in mixed populations, diploids seemed to occur in more xeric habitats, tetraploids in more mesic habitats and triploids either in intermediate ones or growing in very close proximity to either of the two cytotypes. However, Nesom (1983) concluded from analyses of a number of sites throughout the range, that the occurrence of a particular cytotype of G. urceolata is not always correlated with slope, aspect, elevation or accompanying vegetation.
Habitat differentiation between other polyploid species and their diploid progenitors has been demonstrated in a number of field and laboratory studies (Levin, 1983). Recently, Felber-Girard et al. (1996), studying the autopolyploid Anthoxanthum alpinum, demonstrated habitat differentiation in a diploid–tetraploid hybrid zone. Similarly, Bretagnolle & Thompson (1996) found ecological differences between diploid and tetraploid Dactylis glomerata when grown in four contrasting environments. The existence of habitat differentiation between diploids and tetraploids seems likely, but more research on the patterns of intercytotype mating and ecological differences among cytotypes are required to understand the relative importance of these mechanisms to the distribution of G. urceolata.
Triploids
One major finding in this study was the high frequency of triploids (11%) throughout the range of overlap between diploid and tetraploid G. urceolata. A previous study by Nesom (1983) also showed that triploids occurred in G. urceolata, but no estimate of their frequency in populations, nor the extent to which they vary between populations, has been measured. The prevalence of triploids reported in this study is at odds with the idea that triploids are generally inviable as a result of genetic imbalances, and in contrast with much empirical research on autopolyploids, in which triploids were rare, or not found at all (Soltis, 1984; Felber-Girard et al., 1996).
Galax urceolata exhibits clonal growth and therefore it is possible that we have overestimated the number of triploids by repeatedly sampling the same genets. We attempted to maximize the number of genets sampled by positioning sample locations at 2 m intervals. Although some oversampling of genets may still occur, it is unlikely that it has had a major influence on our estimates of cytotype frequency for a number of reasons. First, the physical boundaries of clones were often visible and thus repeated sampling could be avoided. Secondly, when triploids were found, they were often distributed throughout the population, rather than being restricted to one locality. Finally, repeated sampling of some genets would result in a biased estimate only if it occurred disproportionately in one cytotype. However, there is no evidence that clone size and the likelihood of oversampling differed among cytotypes. Collectively, these observations suggest that not only are triploids present in some populations, but they have probably arisen through separate origins in each case.
Triploid G. urceolata occurred in all possible combinations with diploids and tetraploids (i.e. 2x and 3x; 4x and 3x; 2x, 4x and 3x). However, triploids were statistically more likely to occur in populations containing both diploids and tetraploids. This result may reflect the pathway of triploid formation in natural populations through the union of n and 2n gametes. Tetraploids appear to be fertile and most likely produce 2n gametes. This would explain the prevalence of triploids in diploid/tetraploid populations. If diploids are also capable of producing 2n gametes, this may account for the presence of diploid/triploid populations. The existence of triploid/tetraploid populations is more difficult to explain based on commonly accepted pathways of formation (Ramsey & Schemske, 1998). Perhaps these mixtures represent transitional stages towards tetraploidy, in which the diploid parental cytotype has been lost.
The tendency for triploids to occur in the presence of both diploids and tetraploids may suggest as much about factors favouring the establishment of cytotypes as it does the pathways of formation. If triploids are intermediate compared to their diploid and tetraploid parents, they would be most likely to occur in intermediate habitats. These habitats may represent marginal environments where both diploids and tetraploids can also persist. If this were the case, we would expect to see more populations nearly or completely dominated by triploids. The fact that this does not occur suggests that ecological differentiation alone is unlikely to explain the population cytotype structure seen in G. urceolata.
This study suggests that the distribution of cytotypes in G. urceolata is governed by a combination of both genetic and ecological variables. Further examination of the relative fitness of diploids, triploids and tetraploids, as well as patterns of mating and reproductive isolation, is necessary to assess their relative contribution to the maintenance and distribution of polyploid complexes. Further investigation of these issues in mixed cytotype species such as G. urceolata will continue to improve our understanding of polyploid evolution.
References
Arnold, M. L. (1997). Natural Hybridization and Evolution. Oxford University Press, New York.
Arugumanathan, E. and Earle, E. D. (1991). Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol Biol Rep, 9: 229–233.
Baldwin, J. T. Jr. (1941). Galax: The genus and its chromosomes. J Hered, 32: 249–254.
Bino, R. J., De Vries, J. N., Kraak, H. L. and van Pijlen, J. G. (1992). Flow cytometric determination of nuclear replication stages in tomato seeds during priming and germination. Ann Bot, 69: 231–236.
Bretagnolle, F. and Thompson, J. D. (1996). An experimental study of ecological differences in winter growth between diploid and autotetraploid Dactylis glomerata. J Ecol, 84: 343–351.
Costich, D. E., Ortiz, R., Meagher, T. R., Bruederle, L. P. and Vorsa, N. (1993). Determination of ploidy level and nuclear DNA content in blueberry by flow cytometry. Theor Appl Genet, 86: 1001–1006.
Endler, J. A. (1977). Geographic Variation, Speciation and Clines. Princeton University Press, Princeton, NJ.
Epes, D. A. and Soltis, D. E. (1985). An electrophoretic investigation of Galax urceolata (Diapensiaceae). Abstract. Am J Bot, 71: 165–165.
Evans, G. M. (1968). Nuclear changes in flax. Heredity, 23: 25–38.
Felber, F. (1991). Establishment of a tetraploid cytotype in a diploid population: effect of relative fitness of the cytotypes. J Evol Biol, 4: 195–207.
Felber-Girard, M., Felber, F. and Buttler, A. (1996). Habitat differentiation in a narrow hybrid zone between diploid and tetraploid Anthoxanthum alpinum. New Phytol, 133: 531–540.
Fowler, N. L. and Levin, D. A. (1984). Ecological constraints on the establishment of a novel polyploid in competition with its diploid progenitor. Am Nat, 124: 701–711.
Gleason, H. A. (1952). The New Britton and Brown Illustrated Flora of the Northeastern United States and Adjacent Canada. Hafner Press, New York.
Harrison, R. G. and Rand, D. M. (1989). Mosaic hybrid zones and the nature of species boundaries. In: Otte, D. & Endler, J. A. (eds) Speciation and its Consequences, pp. 111–133. Sinauer Associates, Sunderland, MA.
Husband, B. C. and Schemske, D. W. (1998). Cytotype distribution at a diploid–tetraploid contact zone in Chamerion (Epilobium)angustifolium (Onagraceae). Am J Bot, 85: 1688–1694.
Keeler, K. H. (1989). Distribution of polyploid variation in big bluestem (Andropogon gerardii, Poaceae) across the tallgrass prairie region. Genome, 33: 95–100.
Levin, D. A. (1975). Minority cytotype exclusion in local plant populations. Taxon, 24: 35–43.
Levin, D. A. (1983). Polyploidy and novelty in flowering plants. Am Nat, 122: 1–25.
Lewis, W. H. and Suda, Y. (1976). Diploids and polyploids from a single species population: temporal adaptations. J Hered, 67: 391–393.
Masterson, J. (1994). Stomatal size in fossil plants: evidence for polyploidy in the majority of angiosperms. Science, 264: 421–424.
Michaelson, M. J., Price, H. J., Ellison, J. R. and Johnston, J. S. (1991). Comparison of plant DNA contents determined by Feulgen microspectrophotometry and laser flow cytometry. Am J Bot, 78: 183–188.
Nesom, G. L. (1983). Galax (Diapensiaceae): Geographical variation in chromosome number. Syst Bot, 8: 1–14.
Price, H. J. (1988). DNA content variation within angiosperm species. Evol Trends Plants, 2: 53–60.
Ramsey, J. and Schemske, D. W. (1998). Pathways, mechanisms and rates of polyploid formation in flowering plants. Ann Rev Ecol Syst, 29: 467–501.
Soltis, D. E. (1984). Autopolyploidy in Tolmiea menziesii (Saxifragaceae). Am J Bot, 71: 1171–1174.
Soltis, D. E., Bohm, B. A. and Nesom, G. L. (1983). Flavonoid chemistry of cytotypes in Galax (Diapensiaceae). Syst Bot, 8: 15–23.
Song, K., Lu, P., Tang, K. and Osborn, T. C. (1995). Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc Natl Acad Sci USA, 92: 7719–7723.
Stebbins, G. L. (1950). Variation and Evolution in Plants. Columbia University Press, New York.
Stebbins, G. L. (1971). Chromosomal Evolution in Higher Plants. Addison–Wesley, Reading, MA.
Thompson, J. D. and Lumaret, R. (1992). The evolutionary dynamics of polyploid plants: origins, establishment and persistence. Trends Ecol Evol, 7: 302–307.
van Dijk, P. and Bijlsma, R. (1994). Simulations of flowering time displacement between two cytotypes that form inviable hybrids. Heredity, 72: 522–535.
van Dijk, P., Hartog, M. V. and van Delden, W. (1992). Single cytotype areas in autopolyploid Plantago media L. Biol J Linn Soc, 46: 315–331.
Warmke, H. E. (1935). A permanent root tip smear method. Stain Technol, 10: 101–103.
Acknowledgements
We thank P. Briggs, P. Pauls, and A. Sutton for technical assistance, R. Evans, H. Kubiw, T. McDaniel, P. Briggs and A. Sutton for field assistance and H. Kubiw for editorial assistance. We also thank R. Bruce for the use of facilities at Highlands Biological Station, G. Kauffman from the Nantahala National Forest, C. Wentworth from the Chattahoochee–Oconee National Forest, F. Huber from the George Washington and Jefferson National Forests, B. Teague and L. McElrath from the National Park Service – Blue Ridge Parkway, T. Wagoner from Hanging Rock State Park and D. Smith from the Cliffs of the Neuse State Park for allowing the collection of plant samples and providing valuable information. This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada to B.C.H.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Burton, T., Husband, B. Population cytotype structure in the polyploid Galax urceolata (Diapensiaceae). Heredity 82, 381–390 (1999). https://doi.org/10.1038/sj.hdy.6884910
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6884910
Keywords
This article is cited by
-
Cytotype coexistence in the field cannot be explained by inter-cytotype hybridization alone: linking experiments and computer simulations in the sexual species Pilosella echioides (Asteraceae)
BMC Evolutionary Biology (2017)
-
Chromosome numbers, characterization of chromosomal pairing during meiosis, origin and natural propagation in polyploid cytotypes (4x, 5x and 6x) of Agrimonia eupatoria L. (Rosaceae) in northwest Himalayas (India)
Protoplasma (2014)
-
Niche differentiation between diploid and hexaploid Aster amellus
Oecologia (2008)
-
Polyploidy, reproductive biology, and Rosaceae: understanding evolution and making classifications
Plant Systematics and Evolution (2007)