Abstract
The geographic mosaic theory of coevolution is stimulating much new research on interspecific interactions. We provide a guide to the fundamental components of the theory, its processes and main predictions. Our primary objectives are to clarify misconceptions regarding the geographic mosaic theory of coevolution and to describe how empiricists can test the theory rigorously. In particular, we explain why confirming the three main predicted empirical patterns (spatial variation in traits mediating interactions among species, trait mismatching among interacting species and few species-level coevolved traits) does not provide unequivocal support for the theory. We suggest that strong empirical tests of the geographic mosaic theory of coevolution should focus on its underlying processes: coevolutionary hot and cold spots, selection mosaics and trait remixing. We describe these processes and discuss potential ways each can be tested.
Similar content being viewed by others
Introduction
Coevolution among species and the impacts of geography on evolution have always been major research areas within evolutionary biology. However, interest in studying coevolution in a geographic context has recently escalated. A search of the ISI Web of Science shows a dramatic increase over the last 15 years in the number of publications considering both coevolution and geography (Figure 1). Moreover, a growing portion of these papers cite the publications of John N Thompson, whose book ‘The Geographic Mosaic of Coevolution’ (Thompson, 2005) is the latest treatise on this emerging sub-discipline (Figure 1). Thompson's book and papers provide a compendium of the empirical literature on interacting species whose ranges span broad and variable landscapes. They also advance the geographic mosaic theory of coevolution, which is a particular thesis regarding the role of evolutionary and ecological processes in determining patterns of coevolutionary outcome across geographical localities.
Publications that consider coevolution in a geographic context noting those that cite the work of John N Thompson and his geographic mosaic theory of coevolution. Shown are results of an ISS Web of Science Search with criterion ‘(coevolution OR co-evolution) AND geograph*’ sorted by year and divided between articles that cite or do not cite Thompson. Articles with Thompson as an author were excluded from the data. *Projection based on data from the first 9 months of 2006.
The geographic mosaic theory of coevolution (‘GMTC’) hypothesizes that three processes are the primary drivers of coevolutionary dynamics: intermingled coevolutionary hot and cold spots, selection mosaics and trait remixing. For a pair of interacting species, a ‘hot spot’ is a location where the fitness of both species is affected by the distribution of traits in the other species. In contrast, in a ‘cold spot’ the fitness of at least one of the species is unaffected by the other. The term ‘selection mosaic’ refers to variability in the functions that describe reciprocal fitness interactions across space. Finally, ‘trait remixing’ includes gene flow across landscapes, random genetic drift within populations, extinction and recolonization of local populations and mutation. The GMTC predicts that these three processes lead to three observable patterns: spatial variation in the traits mediating an interspecific interaction, trait mismatching among interacting species and few species-level coevolved traits (Thompson, 1999, 2005).
Here, we aim to explain these hypothesized processes and predicted patterns clearly, and highlight empirical approaches to use – and to avoid – when testing the geographic mosaic theory of coevolution. Note that this brief review does not consider the historical context or development of the GMTC; interested readers should consult Thompson (2005).
Verifying predicted patterns is not enough
The geographic mosaic theory of coevolution predicts that hot and cold spots, selection mosaics and trait remixing should result in the three major patterns of coevolution described above. Although mathematical models confirm that these predicted patterns follow from the assumptions of the GMTC (e.g., Gomulkiewicz et al., 2000), identical patterns could be generated without invoking selection mosaics, trait remixing or intermingled coevolutionary hot and cold spots. We illustrate this point using the example of an antagonistic, cyclical interaction, although similar arguments can be developed readily for all major forms of ecological interactions.
Spatially variable traits
The GMTC predicts that traits mediating species interactions should be spatially variable, yet this pattern will almost inevitably be produced by antagonistic coevolution even in the absence of selection mosaics, trait remixing or occasional cold spots. For example, take coevolution between a host and parasite mediated by quantitative traits in each species (Figure 2). In the absence of gene flow, coevolutionary dynamics are likely to be out of phase among patches because of slight variations in initial phenotype frequencies (Morand et al., 1996). Consequently, traits mediating the interaction are likely to vary across space (Figure 2).
Coevolutionary dynamics of trait means, z1 and z2 (left hand panels), and trait mismatching, z1–z2 (right hand panels), for an interaction between a host/prey species (z1; gray lines) and a parasite/predator species (z2; black lines) occurring in two completely isolated populations. The only difference between the two populations labelled a and b is initial allele frequencies at the underlying loci. Dashed vertical lines indicate generation 4000, and highlight the significant difference in trait means that occurs across populations at that time even in the absence of any of the essential processes of the GMTC. Figures were generated based upon the genetically explicit multilocus simulations described in Nuismer et al. (2005).
Sensitivity to historical genotype frequencies also occurs in mutualistic and competitive interactions (Parker, 1999; Nuismer et al., 2000; Thompson et al., 2004). Documenting spatial variation in traits mediating an interaction therefore does not by itself provide clear support for the GMTC over alternative explanations. However, such observations may be critical in identifying traits of importance to an interaction, and may warrant further study in the context of the GMTC.
Trait mismatches
A second major pattern predicted by the GMTC is that the traits mediating an interaction will be well matched in some localities but mismatched in others. This prediction has been evaluated in multiple empirical studies by comparing the extent to which traits mediating an interaction mismatch across multiple locales (Brodie et al., 2002; Zangerl and Berenbaum, 2003; Siepielski and Benkman, 2004). Evaluating whether the results of these studies support the GMTC, however, is made difficult by the absence of any formal definition for a ‘match’ or ‘mismatch’ (Thompson, 1994, 1999, 2005).
An intuitive definition of matching can be made in terms of functional relationships among coevolving traits. For example, pollinators and the flowers they pollinate must emerge at about the same time; the functional relationship between pollinator emergence time and plant flowering time thus defines trait matching. Such relationships have primarily been determined via regression techniques (e.g., Benkman, 1999; Brodie et al., 2002; Brodie and Ridenhour, 2003; Toju and Sota, 2006); the coefficient of determination (R2) then indicates how well matched a pair of traits are across a geographic mosaic. Techniques used in determining allometric scaling relationships (such as reduced major-axis regression; Sokal and Rohlf, 1995) may be particularly useful in determining functional trait matching. Note, however, that in some cases trait matching is not expected. For example, trait values should continually increase when ‘bigger is better’ in the absence of fitness trade-offs (i.e. costs) and thus trait matching should not be evident.
An alternative, indirect approach to evaluating trait mismatching, is to quantify levels of local maladaptation in interacting species (Nuismer, 2006). Local maladaptation has been defined both verbally and mathematically, and has been subject to extensive empirical and theoretical investigation (Gandon et al., 1996; Lively and Jokela, 1996; Morand et al., 1996; Kaltz and Shykoff, 1998; Kaltz et al., 1999; Oppliger et al., 1999; Gandon and Michalakis, 2002; Thrall et al., 2002; Dybdahl and Storfer, 2003; Kawecki and Ebert, 2004; Lively et al., 2004; Nuismer, 2006). Local adaptation has been characterized theoretically within two broad frameworks whose appropriateness is under debate: home versus away and local versus foreign (Kawecki and Ebert, 2004). In contrast, empirical approaches to determining local adaptation involve either reciprocal cross-infection or reciprocal transplant experimental designs. These designs involve measuring and comparing the fitness of individuals within their natal and non-natal environments. Several studies have used such an approach to document local maladaptation in at least one member of an interacting species pair (e.g., Morand et al., 1996; Kaltz and Shykoff, 1998; Kaltz et al., 1999; Oppliger et al., 1999).
Neither trait mismatching nor local maladaptation provide a rigorous test of the GMTC, as both can evolve in the absence of selection mosaics, coevolutionary cold spots or trait remixing. For example, Figure 2 shows there can be strong temporal and spatial variation in the degree of trait mismatching despite lacking the central evolutionary processes invoked by the GMTC. The same is true for local maladaptation which has been demonstrated more rigorously by Morand et al. (1996). Despite this, it is important to emphasize that – at least from a theoretical perspective – selection mosaics and coevolutionary cold spots can greatly increase the likelihood of observing local maladaptation or trait mismatching (Gomulkiewicz et al., 2000; Nuismer et al., 2000, 2003; Nuismer, 2006), although they are not required for such patterns to occur. Thus, these patterns do not provide unequivocal support for the GMTC either.
Few species level coevolved traits
The final major predicted pattern of the GMTC is that few traits involved in an interaction will ultimately become fixed within a pair of species or at higher taxonomic levels. This follows from the first prediction in that if the majority of traits important to an interaction vary across space, then only the few remaining traits that are uniform across space could qualify as being coevolved at the level of the entire species.
Because this prediction is a simple corollary of the first, it too can easily be explained without recourse to the specific processes that make up the GMTC. Again, consider the simple example of antagonistic coevolution (Figure 2). The antagonistic interspecific interactions cause the trait means of the coevolving species to differ across the communities despite the fact that they are identical in all respects with the exception of their initial allele frequencies. Thus, we observe spatial variation in the traits important to the interaction, rather than spatially uniform coevolved traits, simply because the two communities differed in their initial allele frequencies. Similar scenarios can be easily developed for mutualistic and competitive interactions (Parker, 1999; Nuismer et al., 2000).
In summary, its main ecological predictions regarding patterns of coevolution are not unique to the GMTC. This does not invalidate the GMTC. Moreover, future theoretical work may identify additional predicted patterns that prove exclusive to the GMTC. Regardless, the GMTC as it currently stands does advance a specific set of hypotheses about the underlying processes that give rise to Thompson's predicted coevolutionary patterns. Distinguishing a geographic mosaic from alternative evolutionary processes, therefore, requires direct empirical examination of the ecological and population genetic processes that can give rise to spatially variable interactions.
This brings us to the first – and main – set of dos and don'ts for testing the GMTC: don't just verify the predicted patterns. Do test the processes hypothesized to form its foundation.
Processes of the geographic mosaic theory
Because, in our view, the geographic mosaic theory of coevolution is best tested by examining its processes – coevolutionary hot and cold spots, selection mosaics, and trait remixing – we turn to describing each in more detail and discuss ways they can be tested.
Coevolutionary hot and cold spots
The GMTC assumes that fitness interactions among species vary geographically in intensity. In the language of the theory, a region in which reciprocal selection occurs is a ‘coevolutionary hot spot,’ whereas a region where reciprocal selection is absent is termed a ‘coevolutionary cold spot’ because the fitness of at least one species is completely unaffected by the other (Figure 3). Coevolutionary cold spots arise for many reasons, including nonoverlapping geographic ranges (Nuismer et al., 2003), the presence or absence of additional species (Benkman et al., 2001) or shifting abiotic conditions (Hochberg and van Baalen, 1998).
Interspecific frequency-dependent fitness surfaces in cold versus hot spots. Each point on a surface corresponds to the fitness of an individual of one species as a function of its own phenotype and the mean phenotype of the partner species with which it interacts. The thicker lines on each surface indicate the frequency-dependent fitness function for a particular phenotype of one species (left column: species 1 fitness functions; right column: species 2 fitness functions). In cold spots (top row), the fitness function of at least one species does not depend on the mean phenotype of the other species (top right, white line). Fitnesses of both species depend on the other species' mean phenotype in hot spots (bottom row).
In principle, identifying a coevolutionary hot spot is relatively straightforward, and requires demonstrating only that reciprocal selection occurs in a natural population. In practice, however, establishing that reciprocal selection exists in natural populations has proven challenging for numerous reasons. These difficulties include identifying traits that mediate an interaction, measuring fitness consequences of interactions for both species and inferring reciprocal selection from these data in a statistically rigorous fashion.
Largely because of the challenges inherent to establishing reciprocal selection, many empirical studies have relied on comparative spatial analyses of phenotypes or levels of host resistance to identify potential coevolutionary hot spots (Brodie et al., 2002; Zangerl and Berenbaum, 2003; Laine, 2006) rather than measuring reciprocal selection per se. Other empirical studies have directly measured the strength of phenotypic selection, but have measured selection on each of the component species in different geographic regions (Benkman et al., 2003) or on only one of the component species (Rudgers and Strauss, 2004). These studies are important advances in our current understanding of species interactions and they have identified promising empirical approaches (and systems) that should eventually allow full confirmation of the existence of the intermingled coevolutionary hot and cold spots postulated by the GMTC.
One of the most clear-cut approaches that future studies can take to identify coevolutionary hot spots fully and rigorously is to use extensions of the single-species regression approaches pioneered by Lande and Arnold (1983) for estimating selection gradients. Specifically, the recent development of selective source analysis (Brodie and Ridenhour, 2003; Ridenhour, 2005), by which the contribution of reciprocal selection to observed selection gradients can be quantified, should greatly facilitate detection of coevolutionary hot spots. The data required for this statistical framework are (1) traits of interacting individuals and (2) fitness consequences of individual interactions for both component species. Although challenging, these data can plausibly be collected for many types of interactions.
Coevolutionary cold spots can be more challenging to detect than hot spots. This is mainly because when both species are present, it can be difficult to rule out a lack of statistical power in failing to detect reciprocal selection or be confident that target traits in each species have been correctly identified. A significant exception is when a component species is completely absent from a locality.
It is tempting to define hot and cold spots relative to specific traits that might be subject to reciprocal selection in some places but not others. This is unnecessary, however, if one adopts a multivariate view of interacting phenotypes where fitness depends on the complete set of characters contributing to interactions somewhere in the species' joint range. A cold spot then corresponds to the absence of reciprocal multivariate selection relative to the entire suite of traits that could potentially affect interacting species across their collective extent.
To sum up, our dos and don'ts for evaluating hot and cold spots are: do use regression approaches, selective source analysis and multivariate approaches to infer hot spots. When looking to establish cold spots for a particular interaction, don't forget to include regions where one of the species is completely absent.
Selection mosaics
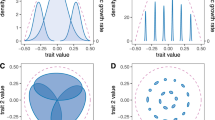
The second – and most distinctive – basic hypothesis of the GMTC is the selection mosaic. ‘Selection mosaic’ refers to spatial variation in the interspecific frequency-dependent fitness functions describing reciprocal selection among interacting species (Thompson, 2005, pp 100–101). To illustrate this concept, consider Figure 4, which shows frequency-dependent fitness surfaces for one of two species that interact across two locations. Each surface shows how the fitness function of species 1 depends on the mean phenotype of species 2 at a particular location. The upper half of the figure shows a case in which the fitness surfaces are the same at the two locations. This is not a selection mosaic. By contrast, the lower part of the figure portrays a scenario in which the fitness surfaces are different in the two locations. This is a selection mosaic.
Local fitness scenarios with and without a selection mosaic. Shown are fitness surfaces of species 1 in two locations A and B with a selection mosaic (bottom row) and without (top row). The white contour on each surface shows the fitness function experienced by species 1 in each locality at a particular time. In the top row, the mean trait of species 2 is 12.5 in population A and 2.5 in B whereas it is 2.5 in both places in the bottom row. The strength of directional selection experienced by species 1 (βspp. 1) is indicated in each locality. The fitness function shown in the top panels and bottom right panel is w(x∣ȳ)=1/{1+exp[(x−ȳ)/4]}, where x is the trait value in species 1 and ȳ is the mean in species 2. The fitness function in the lower left panel is w(x∣ȳ)=exp[−(x−ȳ)2/40]. Values of βspp. 1 were calculated assuming x is normally distributed with mean 7.5 in all panels; the s.d. is 2.5 in all panels except the lower left where it is 2.85. See text for further discussion.
Cold spots represent a distinct form of interspecific frequency-dependent fitness (namely, its absence) and so it may seem natural to treat them as being part of a selection mosaic. However, Thompson's original definition of a selection mosaic refers strictly to populations where reciprocal selection occurs. Thus, his definition specifically excludes cold spots.
A selection mosaic, as formally defined, is not equivalent to spatial variation in the strength or direction of reciprocal selection. The difference is subtle and can be easily missed. Indeed, spatially variable or spatially uniform selection can occur in the presence or absence of a selection mosaic. To see this, consider the upper scenario in Figure 4, which is not a selection mosaic because the fitness surfaces are the same. If the mean phenotype of species 2 differs between the two habitats, then species 1 would experience spatially variable selection in the absence of a selection mosaic (compare the fitness functions realized by species 1 in the two locations – represented by the two white contours – and the resulting strengths of local directional selection). It is theoretically possible, but exceedingly improbable in practice, that a species can experience geographically uniform selection in a selection mosaic if the distributions of the interacting species vary in a specific way among sites. The lower portion of Figure 4 shows such an exception: this is a selection mosaic because the fitness surfaces at the two locations are different yet species 1 experiences spatially uniform directional selection.
As a single frequency-dependent fitness function can be consistent with spatially variable selection when phenotypic distributions vary across space (Figure 4, top), it is important to show empirically that fitness functions differ in different locations to document a selection mosaic. One approach is to use the fact that reciprocal selection can vary across a landscape over which phenotypic distributions of interacting species are spatially uniform only if that landscape is a selection mosaic.
Testing for selection mosaics thus depends both on detecting reciprocal selection at different localities (i.e., identifying multiple coevolutionary hot spots) and on establishing variability in frequency-dependent fitness functions across these same localities. A bona fide selection mosaic, as specifically defined by Thompson's theory (op. cit.), has yet to be rigorously identified for any coevolutionary system. Nevertheless, several empirical studies have used either clever experimental manipulations (e.g., Rudgers and Strauss, 2004) or surveys of geographical variation in traits combined with measurements of phenotypic selection (e.g., Benkman, 1999; Benkman et al., 2003) to suggest their existence.
An important challenge for future empirical and theoretical work will be developing novel experimental designs and statistical tools that can rigorously test for variability in frequency-dependent fitness functions. Probably, the most direct way to detect a selection mosaic empirically is to carefully replicate genotype frequencies in all species across different localities. If reciprocal selection varies across sites for these replicated communities, a selection mosaic must be present. (The absence of such variation is inconclusive.) Alternatively, one may be able to develop statistical methods that control for different underlying genotypic distributions in selective source analysis (Brodie and Ridenhour, 2003; Ridenhour, 2005). And as interaction fitnesses are essentially functions of partner species' phenotypes, another approach might be to apply function-valued trait methods (Kirkpatrick and Heckman, 1989; Kingsolver et al., 2001) to identify selection mosaics by statistically distinguishing different functional forms of reciprocal selection.
In summary: don't confuse selection mosaics with variable selection; they are not the same. Do investigate whether frequency-dependent fitness functions – and not just the strength of selection – vary across space.
Trait remixing
Although selection mosaics await full empirical confirmation, the third premise of the GMTC, trait remixing, represents ‘the uncharted waters of coevolutionary research’ (Thompson, 2005). Nevertheless, trait remixing is the direct basis for the GMTC's prediction that interacting species are sometimes mismatched or locally maladapted. Trait remixing tends to produce locally mismatched phenotypes or local maladaptation because it continually alters the spatial distributions of alleles and traits that mediate interspecific interactions thereby interfering with local selection. There are four potential mechanisms, which we discuss in turn: gene flow across landscapes, random genetic drift within populations, extinction and recolonization of local populations, and mutation.
Under the GMTC, gene flow is sometimes considered a source of local maladaptation and trait mismatching because traits or alleles shaped by selection in one community context are introduced into a different context (Nuismer et al., 1999; Gomulkiewicz et al., 2000). Mismatching can be common among populations; in parsnip webworms, phenotypic mismatch occurred in eight of 20 populations, and the best explanatory variable was proximity to populations on an alternate host, suggesting the importance of migration (Zangerl and Berenbaum, 2003).
However, gene flow can also be a creative force, providing genetic variation for reciprocal selection and continued coadaptation. For example, under antagonistic coevolution, theoretical studies show that, provided migration rates are low, the species with the relatively greater amount of gene flow will generally show adaptation, rather than maladaptation (Gandon et al., 1996; Gandon, 2002). A number of empirical studies of hosts and parasites have examined local adaptation and relative gene flow and support this theoretical result (reviewed in Dybdahl and Storfer, 2003).
Whether gene flow is an important source of trait remixing depends on the relative geographic scales and strength of selection mosaics compared to gene flow. If dispersal distances are small compared to the sizes of selectively distinct patches, then gene flow should play a minor role in trait remixing. When dispersal distances span selectively different patches, the effect of gene flow will depend on the heterogeneity of selection across the selection mosaic. When selective differences are weak among patches, gene flow has a small effect on trait remixing. However, gene flow might be an important influence when gene flow is high and selective differences are large across the mosaic. Thus, testing the importance of gene flow in trait remixing requires estimating gene flow (e.g. using neutral genetic markers) across the same landscape for which the scale and strength of selection mosaics have been mapped. Although no study has yet mapped a selection mosaic (see above), gene flow and the spatial genetic structures of interacting species pairs have been estimated using neutral genetic markers (e.g., Mulvey et al., 1991; Dybdahl and Lively, 1996; Althoff and Thompson, 1999; McCoy et al., 2005; Prugnolle et al., 2005; review in Criscione and Blouin, 2006).
Mapping the geographic scale of gene flow relative to selection mosaics requires sampling genetic marker variation at multiple sites across the (putative) selection mosaic. The importance of gene flow in trait remixing would be indicated by a relatively large proportion of genetic variation partitioned across selectively distinct patches (AMOVA, Schneider et al., 2000), or the identification of genetically connected populations that extend across selectively distinct patches (STRUCTURE, Pritchard et al., 2000). Assignment methods (Cornuet et al., 1999; Manel et al., 2005) might offer the possibility of determining whether a particular population receives immigrants from populations subject to a different selection regime.
A second trait remixing process, random genetic drift, might lead to trait remixing if the force of selection is weaker than drift. If so, alleles that underlie traits may drift to high frequencies despite selection. If predominant trait values within populations result from drift rather than selection, then levels of divergence among populations of traits that mediate interspecific interactions should resemble levels observed in selectively neutral traits.
Genetic marker variation can provide a neutral expectation against which the landscape pattern of trait variation can be compared (Spitze, 1993; reviewed in Merila and Crnokrak, 2001; McKay and Latta, 2002). One approach is to compare the phylogeographies of species of interest with other independent species across the same landscape. Incongruent phylogeographies could suggest that selection rather than drift (or other historical factors) drove trait variation (Thompson and Calsbeek, 2005).
Another means of distinguishing adaptive and neutral causes of geographical variation is to compare relative levels of among-population divergence in quantitative traits and neutral genetic markers. For example, levels of among-population divergence for neutral traits, driven by drift and migration only, can be estimated with FST. At the same time, levels of phenotypic divergence in coevolving traits, driven by drift, migration and selection, can be estimated with QST. Drift could be ruled out as a plausible explanation of the observed divergence in coevolving traits if FST << QST. It is also possible to incorporate information on geographic distances among patches in a selection mosaic following Storz (2002), where both FST and QST should be correlated with geographical distance if the distribution of coevolving traits is governed by drift rather than selection. Such comparisons are most robust for phenotypic traits with a predominantly additive basis in outbred species (López-Fanjul et al., 2003; Goudet and Büchi, 2006).
Metapopulation dynamics, driven by the extinction and recolonization of populations, can also alter the spatial distribution of alleles created by gene flow and drift (Slatkin, 1987) and thereby provide yet another mechanism for trait remixing. According to theory, when recolonization of extinct populations follows the ‘propagule pool’ model (colonists originate from a subset of populations), neutral variation among populations can be greater than migration-drift equilibrium expectations without extinction/recolonization. In contrast, ‘migrant pool’ recolonization (colonists originate from the entire set of populations) leads to relative homogenization of neutral genetic variation among populations (Slatkin, 1987; Whitlock and McCauley, 1990). Thus metapopulation dynamics can, depending on the mode of colonization, have opposite effects on spatial genetic structure.
In the context of coevolution, metapopulation dynamics can drive the pattern and geographic scale of local adaptation and maladaptation between interacting species, leading to local adaptation either among populations within a metapopulation (Thrall et al., 2002) or among metapopulations (Laine, 2005). Understanding the importance of metapopulation dynamics in a coevolutionary system requires longitudinal studies or occupancy surveys of a large number of patches to estimate the rate of population extinction and recolonization and detailed studies of the source populations of colonists (Whitlock, 1992; Dybdahl, 1994; Ingvarsson et al., 1997).
Mutation is the ultimate source of new traits and alleles, and also a potential cause of trait remixing. But detecting new mutations and their fitness consequences in natural populations – particularly for traits important to an interspecific interaction – is difficult. Nevertheless, experimental coevolutionary studies with bacteria and phage have shown that mutation can be the source of variation in coevolutionary trajectories among replicate populations (Buckling and Rainey, 2002).
Here, then, are some dos and don'ts for testing trait remixing. Don't measure gene flow without comparing it to the scale and strength of selection mosaics. Do compare the spatial structures of interacting phenotypes and neutral genetic markers to assess the importance of gene flow, local selection and random genetic drift. Although it may be more challenging, do try to measure extinction/recolonization dynamics and spontaneous mutation.
Conclusion
Coevolutionary studies that incorporate geographic structure have contributed substantially to our understanding of species interactions on both ecological and evolutionary time scales. Many of these studies have been inspired by the GMTC. The continuing influence of the GMTC, however, will depend on its verification by strong empirical tests.
We have argued that truly rigorous tests of the GMTC must focus on the hypothesized evolutionary and ecological processes. Unfortunately, testing appropriately for these processes within any empirical system is not easy. Doing so requires careful and extensive documentation of reciprocal and non-reciprocal selection, the forms of selection across the range of the interaction, and trait remixing through gene flow, drift, metapopulation dynamics and mutation. Moreover, appropriate statistical frameworks for testing some aspects of the GMTC have yet to be developed. Many researchers may therefore be discouraged by our recommendations; given the logistical constraints of most empirical systems, it may seem impractical – or even unproductive – to attempt to comprehensively test for a geographic mosaic. We contend, however, that rigorous tests of the GMTC are not only possible but have tremendous potential to increase our practical understanding of coevolution. We propose that testing of the theory can be streamlined using a stepwise, ‘triage’ approach (Figure 5). This strategy, which includes steps similar to ones proposed by Thompson (2005, chapter 8), should at least partly be feasible for even complex or little-known study systems, where practical and logistical hurdles can be high.
The indispensable first step is to gather information on the distribution and natural history of interacting partners within a coevolutionary system. This step could include identifying candidate traits that may be under reciprocal selection, as well as potential coevolutionary hotspots and other physical and biotic factors that might affect the interaction. Second, researchers should gather data that could be used to test for reciprocal selection between coevolving partners using the regression approaches cited above. Of course, one can never completely rule out reciprocal selection as it is always possible that selection might be acting on cryptic traits or in narrowly distributed (or unsampled) coevolutionary hotspots, or that sample sizes are simply too small to detect weak selection. However, until there is a positive evidence of reciprocal selection acting on some traits, in at least some populations, further testing of a geographic mosaic is unnecessary.
If reciprocal selection is present, researchers could next direct their efforts in either of two directions (Figure 5). On one hand, they could measure the spatial genetic structures of the interacting species to determine whether gene flow, drift, extinction/recolonization, or mutation have the potential to produce trait remixing. On the other hand, a researcher could search for cold spots and selection mosaics, perhaps suggested by geographic variation in the physical or biotic environment. Of these two paths, it is probably easiest to test for trait remixing via gene flow or drift and look for cold spots in which one of the interacting species is absent. Regardless of how one proceeds initially, tests of both processes will be needed to fully discern whether the components of trait remixing and divergent selection occur on comparable scales.
Although this final stage presents the most challenging empirical hurdle for testing the geographic mosaic theory, and reaching this stage may require many years of work by multiple investigators, the investigative process itself has great potential to reveal significant knowledge about coevolution. Within natural systems, rigorously testing the evolutionary processes underlying a geographic mosaic can reveal how selection and gene flow act in concert to maintain the diversity of coevolutionary systems over time, even in the face of rapidly changing environments and ecological communities. Additionally, explicit tests of the fundamental processes of the geographic mosaic hold excellent promise to improve our understanding of coevolution in applied settings, such as epidemiology. Indeed, the emergence, spread, and evolution of human infectious diseases can be highly dependent on geographic structuring of populations, communities and selection. Arthropod-vectored diseases such as malaria and Dengue fever, where coevolutionary changes occur extremely rapidly, are two prominent examples in this regard. Information gleaned from our triage approach (Figure 5) or other empirical tests of the GMTC in this context could lead to approaches for combating these diseases (Woolhouse, 2002).
To conclude, do pursue rigorous empirical examinations of the GMTC and don't be put off by the challenges. By focusing research efforts on appropriate and powerful studies of its underlying ecological and evolutionary processes, tests of the geographic mosaic theory of coevolution will illuminate the inner workings of the coevolutionary process.
References
Althoff DM, Thompson JN (1999). Comparative geographic structures of two parasitoid-host interactions. Evolution 53: 818–825.
Benkman CW (1999). The selection mosaic and diversifying coevolution between crossbills and lodgepole pines. Am Nat 153: S75–S91.
Benkman CW, Holimon WC, Smith JW (2001). The influence of a competitor on the geographic mosaic of coevolution between crossbills and lodgepole pine. Evolution 55: 282–294.
Benkman CW, Parchman TL, Favis A, Siepielski AM (2003). Reciprocal selection causes a coevolutionary arms race between crossbills and lodgepole pine. Am Nat 162: 182–194.
Brodie III ED, Ridenhour BJ (2003). Reciprocal selection at the phenotypic interface of coevolution. Integrative and Comparative Biology 43: 408–418.
Brodie III ED, Ridenhour BJ, Brodie Jr ED (2002). The evolutionary response of predators to dangerous prey: hotspots and coldspots in the geographic mosaic of coevolution between garter snakes and newts. Evolution 56: 2067–2082.
Buckling A, Rainey PB (2002). Antagonistic coevolution between a bacterium and a bacteriophage. Proc R Soc B-Biol Sci 269: 931–936.
Cornuet JM, Piry S, Luikart G, Estoup A, Solignac M (1999). New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 153: 1989–2000.
Criscione CD, Blouin MS (2006). Minimal selfing, few clones, and no among-host genetic structure in a hermaphroditic parasite with asexual larval propagation. Evolution 60: 553–562.
Dybdahl MF (1994). Extinction, recolonization, and the genetic structure of tidepool copepod population. Evol Ecol 8: 113–124.
Dybdahl MF, Lively CM (1996). The geography of coevolution: comparative population structures for a snail and its trematode parasite. Evolution 50: 2264–2275.
Dybdahl MF, Storfer A (2003). Parasite local adaptation: Red Queen versus Suicide King. Tr Ecol Evol 18: 523–530.
Gandon S (2002). Local adaptation and the geometry of host–parasite coevolution. Ecol Lett 5: 246–256.
Gandon S, Capowiez Y, Dubois Y, Michalakis Y, Olivieri I (1996). Local adaptation and gene-for-gene coevolution in a metapopulation model. Proc R Soc Lond B 263: 1003–1009.
Gandon S, Michalakis Y (2002). Local adaptation, evolutionary potential and host-parasite coevolution: interactions between migration, mutation, population size and generation time. J Evol Biol 15: 451–462.
Gomulkiewicz R, Thompson JN, Holt RD, Nuismer SL, Hochberg ME (2000). Hot spots, cold spots, and the geographic mosaic theory of coevolution. Am Nat 156: 156–174.
Goudet J, Büchi L (2006). The effects of dominance, regular inbreeding and sampling design on Qst, an estimator of population differentiation for quantitative traits. Genetics 172: 1337–1347.
Hochberg ME, van Baalen M (1998). Antagonistic coevolution over productivity gradients. Am Nat 152: 620–634.
Ingvarsson PK, Olsson K, Ericson L (1997). Extinction-recolonization dynamics in the mycophagous beetle Phalacrus substriatus. Evolution 51: 187–195.
Kaltz O, Gandon S, Michalakis Y, Shykoff JA (1999). Local maladaptation in the anther-smut fungus Microbotryum violaceum to its host plant Silene latifolia: evidence from a cross-inoculation experiment. Evolution 53: 395–407.
Kaltz O, Shykoff JA (1998). Local adaptation in host-parasite systems. Heredity 81: 361–370.
Kawecki TJ, Ebert D (2004). Conceptual issues in local adaptation. Ecol. Lett. 7: 1225–1241.
Kingsolver J, Gomulkiewicz R, Carter P (2001). Variation, selection and evolution of function-valued traits. Genetica 112/113: 87–104.
Kirkpatrick M, Heckman N (1989). A quantitative genetic model for growth, shape, reaction norms, and other infinite-dimensional characters. J Math Biol. 27: 429–450.
Laine AL (2005). Spatial scale of local adaptation in a plant-pathogen metapopulation. J Evol Biol 18: 930–938.
Laine AL (2006). Evolution of host resistance: looking for coevolutionary hotspots at small spatial scales. Proc R Soc B-Biol Sci 273: 267–273.
Lande R, Arnold SJ (1983). The measurement of selection on correlated characters. Evolution 37: 1210–1226.
Lively CM, Dybdahl MF, Jokela J, Osnas EE, Delph LE (2004). Host sex and local adaptation by parasites in a snail-trematode interaction. Am Nat 164: S6–S18.
Lively CM, Jokela J (1996). Clinal variation for local adaptation in a host-parasite interaction. Proc R Soc London Ser B-Biol Sci 263: 891–897.
López-Fanjul C, Fernández A, Toro MA (2003). The effect of neutral nonadditive gene action on the quantitative index of population divergence. Genetics 164: 1627–1633.
Manel S, Gaggiotti OE, Waples RS (2005). Assignment methods: matching biological questions with appropriate techniques. Tr Ecol Evol 20: 136–142.
McCoy KD, Chapuis E, Tirard C, Boulinier T, Michalakis Y, Le Bohec C et al. (2005). Recurrent evolution of host-specialized races in a globally distributed parasite. Proc R Soc B-Biol Sci 272: 2389–2395.
McKay JM, Latta RG (2002). Adaptive population divergence: markers, QTL, and traits. Tr Ecol Evol 17: 285–291.
Merila J, Crnokrak P (2001). Comparison of genetic differentiation at marker loci and quantitative traits. J Evol Biol 14: 892–903.
Morand S, Manning SD, Woolhouse MEJ (1996). Parasite–host coevolution and geographic patterns of parasite infectivity and host susceptibility. Proc R Soc Lond B 263: 119–128.
Mulvey M, Aho JM, Lydeard C, Leberg PL, Smith MH (1991). Comparative population genetic structure of a parasite (Fascioloides magna) and its definitive host. Evolution 45: 1628–1640.
Nuismer SL (2006). Parasite local adaptation in a geographic mosaic. Evolution 60: 24–30.
Nuismer SL, Thompson JN, Gomulkiewicz R (1999). Gene flow and geographically structured coevolution. Proc R Soc London Ser B-Biol Sci 266: 605–609.
Nuismer SL, Thompson JN, Gomulkiewicz R (2000). Coevolutionary clines across selection mosaics. Evolution 54: 1102–1115.
Nuismer SL, Thompson JN, Gomulkiewicz R (2003). Coevolution between hosts and parasites with partially overlapping geographic ranges. J Evol Biol 16: 1337–1345.
Oppliger A, Vernet R, Baez M (1999). Parasite local maladaptation in the Canarian lizard Gallotia galloti (Reptilia: Lacertidae) parasitized by haemogregarian blood parasite. J Evol Biol 12: 951–955.
Parker MA (1999). Mutualism in metapopulations of legumes and rhizobia. Am Nat 153: S48–S60.
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Prugnolle F, Theron A, Pointier JP, Jabbour-Zahab R, Jarne P, Durand P et al. (2005). Dispersal in a parasitic worm and its two hosts: consequence for local adaptation. Evolution 59: 296–303.
Ridenhour BJ (2005). Identification of selective sources: partitioning selection based on interactions. Am Nat 166: 12–25.
Rudgers JA, Strauss SY (2004). A selection mosaic in the facultative mutualism between ants and wild cotton. Proc R Soc London Ser B-Biol Sci 271: 2481–2488.
Schneider S, Roessli D, Excoffier L (2000). ARLEQUIN ver 2.000: A Software for Population Genetics Data Analysis. Genetics and Biometry Laboratory, University of Geneva: Geneva, Switzerland.
Siepielski AM, Benkman CW (2004). Interactions among moths, crossbills, squirrels and lodgepole pine in a geographic selection mosaic. Evolution 58: 95–101.
Slatkin M (1987). Gene flow and the geographic structure of natural populations. Science 236: 787–792.
Sokal RR, Rohlf FJ (1995). Biometry: The Principles and Practice of Statistics in Biological Research. W.H. Freeman and Company: New York.
Spitze K (1993). Population structure in Daphnia obtusa: quantitative genetic and allozymic variation. Genetics 135: 367–374.
Storz JF (2002). Contrasting patterns of divergence in quantitative traits and neutral DNA markers: analysis of clinal variation. Mol Ecol 11: 2537–2551.
Thompson JN (1994). The Coevolutionary Process. University of Chicago Press: Chicago, IL, USA.
Thompson JN (1999). Specific hypotheses on the geographic mosaic of coevolution. Am Nat 153: S1–S14.
Thompson JN (2005). The Geographic Mosaic of Coevolution. University of Chicago Press: Chicago, IL, USA.
Thompson JN, Calsbeek R (2005). Molecular and ecological differentiation of species and species interactions across large geographic regions: California and the Pacific Northwest. In: Fellowes MDE, Holloway GJ, Rolff J (eds). Insect Evolutionary Ecology. Royal Entomological Society: London.
Thompson JN, Nuismer SL, Merg K (2004). Plant polyploidy and the evolutionary ecology of plant/animal interactions. Biol J Linn Soc 82: 511–519.
Thrall PH, Burdon JJ, Bever JD (2002). Local adaptation in the Linum marginale–Melampsora lini host–pathogen interaction. Evolution 56: 1340–1351.
Toju H, Sota T (2006). Imbalance of predator and prey armament: geographic clines in phenotypic interface and natural selection. Am Nat 167: 105–117.
Whitlock MC (1992). Nonequilibrium population structure in forked fungus beetles: extinction, colonization, and the genetic variance among populations. Am Nat 139: 952–970.
Whitlock MC, McCauley DE (1990). Some population genetic consequences of colony formation and extinction: genetic correlations within founding groups. Evolution 44: 1717–1724.
Woolhouse ME (2002). Population biology of emerging and re-emerging pathogens. Trends Microbiol 10: S3–S7.
Zangerl AR, Berenbaum MR (2003). Phenotype matching in wild parsnip and parsnip webworms: causes and consequences. Evolution 57: 806–815.
Acknowledgements
We thank John N Thompson for generously spending part of his field season to discuss the GMTC with us and for his helpful comments on an early draft. We are grateful to Butch Brodie and an anonymous reviewer for providing constructive comments. The National Science Foundation (grants DEB 0209916 and DMS 0540524 to RG, DEB 0343023 and DMS 0540392 to SLN, DEB 0296049 to MFD, and DEB 0516841 to O Pellmyr) and the Natural Sciences and Engineering Research Council of Canada (to WG) provided financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gomulkiewicz, R., Drown, D., Dybdahl, M. et al. Dos and don'ts of testing the geographic mosaic theory of coevolution. Heredity 98, 249–258 (2007). https://doi.org/10.1038/sj.hdy.6800949
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800949
Keywords
This article is cited by
-
Evidence for an innate basis of prey preference in a desert ambush predator
Evolutionary Ecology (2023)
-
Fixed prey cue preferences among Dusky Pigmy Rattlesnakes (Sistrurus miliarius barbouri) raised on different long-term diets
Evolutionary Ecology (2016)
-
The potential of a population genomics approach to analyse geographic mosaics of plant--insect coevolution
Evolutionary Ecology (2011)
-
Weevils and camellias in a Darwin's race: model system for the study of eco‐evolutionary interactions between species
Ecological Research (2011)
-
Arms Race Coevolution: The Local and Geographical Structure of a Host–Parasite Interaction
Evolution: Education and Outreach (2010)