Abstract
Y chromosome variation in 457 Croatian samples was studied using 16 SNPs/indel and eight STR loci. High frequency of haplogroup I in Croatian populations and the phylogeographic pattern in its background STR diversity over Europe make Adriatic coast one likely source of the recolonization of Europe following the Last Glacial Maximum. The higher frequency of I in the southern island populations is contrasted with higher frequency of group R1a chromosomes in the northern island of Krk and in the mainland. R1a frequency, while low in Greeks and Albanians, is highest in Polish, Ukrainian and Russian populations and could be a sign of the Slavic impact in the Balkan region. Haplogroups J, G and E that can be related to the spread of farming characterize the minor part (12.5%) of the Croatian paternal lineages. In one of the southern island (Hvar) populations, we found a relatively high frequency (14%) of lineages belonging to P*(xM173) cluster, which is unusual for European populations. Interestingly, the same population also harbored mitochondrial haplogroup F that is virtually absent in European populations – indicating a connection with Central Asian populations, possibly the Avars.
Similar content being viewed by others
Introduction
Polymorphic markers (biallelic loci and STRs) positioned in the nonrecombining part of Y chromosome (NRY) and paternal inheritance make this chromosome a powerful tool for studying population genetics and evolution.1,2 The use of Y chromosome is increasing as new polymorphisms suitable for the studies of human evolutionary history emerge, mostly because of advanced experimental techniques.3,4 A four times smaller effective population size in comparison with nuclear autosomal loci is reflected in reduced diversity and increased susceptibility to genetic drift.5,6 Therefore, Y chromosome studies are useful in studies of large-scale evolutionary events in human history and population genetics phenomena in small isolated communities.
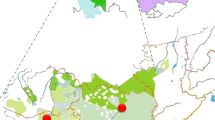
In this paper we use 16 Y SNPs/indel and eight STRs to analyze the diversity in the Croatian mainland and four island populations (Krk, Brač, Hvar and Korčula) (Figure 1) to address three specific problems: (1) How do the Croatian Y chromosomal data fit in the context of other European populations in relation to current understanding of the peopling of the Adriatic (Mediterranean) region? (2) How did long-term isolation and genetic drift affect Y chromosome markers distribution in Croatian islands as compared to the mainland population? (3) How well do these results reflect the extent of isolation and reduced genetic diversity implied by previous anthropological studies?7,8,9,10,11,12,13,14,15,16
Population history
The area under study was continuously inhabited since middle Palaeolithic and is rich in archaeological findings that prove evidence of Mousterian, Aurignacien and Gravettien cultures.17 This, as well as two main episodes of Holocene, the spread of farming and Indo-European languages in Europe, might be important for understanding genetic history of the region. The oldest western Balkan population of a clearly Indo-European origin and a known name is the Illyrian who remained the basic population after which other populations followed such as Greeks, Romans, Avars and Slavs. By the early 6th century AD Croats (Slavs) settled in the region that is today's continental Croatia and in the 6th and 7th century AD they reached the Adriatic Sea coast where they became a predominant population.
The history of the islands' settling was similar to the mainland, but with differences in early farming spreading. Hunting–gathering people inhabited most of the Eastern Adriatic islands, which at the time were still a part of the mainland (20–10 000 years BP). The Adriatic Sea in its current shape was formed relatively recently, about 8–6000 years ago. Before that, its north-west part (including the islands) was a steppe surrounded by the Dinarides, Alpes and Apennines (Figure 1).18,19 The dates and distribution of Impresso pottery, generally considered to appear together with early farming20 along the central and western Mediterranean coast suggests that peopling occurred primarily from the sea. Müller21 proposes that the early farmers arrived from the Apulia area, southern Italy, and that the immigrants–farmers along the coast lived together with native hunter–gatherers in the hinterland for several centuries. An alternative scenario of colonization of the Mediterranean (Adriatic) basin is by land, from the area of the Middle-East, along the southern side of the Dinarides through the steppe which later became the northern Adriatic Sea, to present Italy. In the 4th century BC the Greek colonists organized the colonies on the southern Adriatic islands – Hvar and Korčula and neighboring coastal area. Also, in the Middle ages islands were under the rule of Venice. The ancient Silk Road from China through Central Asia to the Mediterranean could have further contributed to the genetic structure of the investigated populations. Finally, during the Ottoman expansion between the 16th and 18th century AD small successive immigrations to the islands occurred from the Balkans mainland.
Sample and methods
Sample
Blood samples were obtained from 457 Croat volunteers with appropriate informed consent and with the approval of the authorized Ethics Committee. A total of 109 individuals were sampled from six geographically dispersed counties encompassing different regions of Croatia named in this study as Croatian mainland sample while 348 individuals were sampled from four Adriatic islands (Figure 1). A detailed description of sampling procedure for 74 examinees from the island of Krk, 49 from Brač, 91 from Hvar and 134 from Korčula was reported in our previous paper.16 All field studies were preformed by the staff of the Institute for Anthropological Research, Zagreb. The Y chromosomal analyses were performed by LB and MP in the Estonian Biocentre, Tartu, Estonia.
Y chromosome analysis
DNA was extracted from the whole blood according to the macromethod described by Poncz et al.22 The choice of investigated biallelic markers in this study was based on their informativeness in defining haplogroups in this geographic region. The following biallelic markers were typed: M93 was typed according to the method of Hurles et al,23 YAP24 according to Hammer and Horai,25 SRY153226 according to Kwok et al27 (SRY1532 is equivalent to SRY1083128), 92R729 according to Hurles et al,30 SRY8299/406426 according to Santos et al,31 12f232 according to Rosser et al,33 Tat according to Zerjal et al,34 M20, M52, M70, M89, M124, M170, M201, M173 and M175 according to the method of Underhill et al.4 The deep-rooting marker M9 was typed on all samples and other markers were typed hierarchically according to their known phylogeny. The haplogroups were labeled according to the YCC nomenclature.35 A tentative assignment of all our R1 chromosomes, that are derived at M173 but do not have the G to A back mutation at SRY10831, into haplogroup R1b is based on the observations of Cruciani et al.36 Their work supports the monophyletic relatedness of these lineages based on the finding that all of the investigated European (102) and Middle Eastern (8) chromosomes belonging to Jobling's haplogroup 1 shared an M269 mutation.
Two trinucleotide STRs (DYS388 and DYS392) and six tetranucleotide STRs (DYS19, DYS389I, DYS389II, DYS390, DYS391 and DYS393) were typed on all 457 chromosomes. PCR reactions were carried out in the thermocycler ‘Biometra UNO II’ and performed in a 20 μl reaction volume containing 100 ng genomic DNA, 2.5 mM MgCl2, 75 mM Tris-HCl (pH 9.0), 100 μ M of each dNTP, 0.1 μ M of each primer and 2 U Taq DNA polymerase. The PCR cycling conditions used were those described by deKnijf et al.37 PCR products were resolved and detected on 6% denaturing ReproGelTM High Resolution gels (Amersham Pharmacia Biotech) using the Automatic Laser Fluorescence ExpressTM DNA sequencer (Pharmacia Biotech AB). The Allele Links version 1.00 software (Pharmacia Biotech) was used to analyze fragment sizes. The alleles were named according to the number of repeat units, which were established with sequenced allele ladders as suggested by deKnijff et al.37
Allelic frequencies were estimated by the allele counting method. Haplotype diversity within population was calculated using the Arlequin 2.0 package,38 according to the formula

where n is the number of gene copies, k is the number of haplotypes and pi is the sample frequency of the ith haplotype. 95% ‘credible regions’ were calculated from posterior distribution of the proportion of a group of lineages in the population, as in Richards et al.39 Median-joining networks (MJs) were constructed with the program Network 2.0.40
Results
In Table 1, the frequencies of the nine different haplogroups defined in 457 Croatian males divided into five subpopulations are compared. Haplogroup I, defined by M170, is the prevailing Y chromosome group through Croatian mainland and islands with the highest frequency (49% on average) reported in Europe so far.
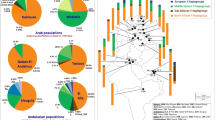
This haplogroup is spread in all European populations with variable frequencies, occuring at >40% in Saami, and at <10% in south (Greeks, Italians) and at ∼20% in west Europe (Dutch, French).33,41 Haplogroup I occurs significantly more frequently in three southern islands – Brač, Hvar and Korčula than in the mainland and Krk in the north (χ2=25.62, 1 d.f., P=0.000). When mainland sample is subdivided by geographic areas higher frequency of I is observed in eastern and southern parts of Croatia while being also the most frequent haplogroup in Central/Western Balkan (Bosnia and Herzegovina – our unpublished data). Although the samples from Yugoslavia were not typed for M170, Semino et al.41 data indicate that most if not all of the unclassified derivates of M89 mutation in Europe are expected to have either M170 or M201 mutation. STR analyses of Croatian haplogroup I-M170 lineages defined 110 haplotypes, 32 of which occurred more than once. The most frequent haplotype, typed by eight STR loci, is 16–13–10–28–24–11–11–13 observed in 31 individuals. A phylogenetic reconstruction of haplogroup I haplotypes occurring three or more times is shown in Figure 2. Many of them are shared between populations because of either parallel mutation and deep time depth or a common origin from a relatively large number of founder haplotypes.
YCC phylogenetic tree, frequencies and phylogenetic relationships. (a) Haplogroup frequencies of the total Croatian sample are indicated in italic below the corresponding branch of the YCC tree. (b) Median joining network of haplogroup R1a and I haplotypes that occurred 3 or more times. The area of each circle is proportional to the frequency of the haplotype. Branch lengths are proportional to the number of single mutational steps.
Most Croatian haplogroup I chromosomes (126/221) share a similar STR haplotype from here on called the Dinaric Modal Haplotype (DMH: 16–24–11–11–13, defined by DYS19–390–391–392–393, respectively) or its one-step derivatives. Interestingly, DMH was not present among 102 Nordic Jobling's haplogroup 2 chromosomes (corresponds to haplogroup I42) reported by Helgason et al.43 Instead, the North Europeans share another modal haplotype 14–23–10–11–13 which is three steps away from the DMH and together with its one step derivatives covers again more than half of haplogroup 2 chromosomes (54/102) in the region of its spread.
Haplogroups R1a and R1b showed the opposite frequency distribution to I-M170 and were more common in the northwest among the mainland population and in the northern island Krk. The frequency of R1b in Croatian sample (7.9%) is comparable to some other Balkan populations, for example, Macedonian (10%),41 Greek (11%) and Yugoslavian (11%),33 but it is considerably lower than in western European populations where its frequency ranges between 40% in Germans and 81% in Irish.33 Unusual for European populations,41,44 14% of Y chromosomes from Hvar carried haplogroup P* without M173 mutation associated with a relatively high number of repeats (n=15) in the DYS392 locus. A detailed description of the occurrence of this haplogroup will be reported elsewhere.
In contrast to R1b, its sister clade R1a shows eastern distribution in Europe being as low as 10–12% in neighboring Balkan populations, Greeks and Albanians, lower in Western Europe, in Italians, Germans and the Dutch (4–6%), but as high as 50–60% in eastern and northern Europe, for example, Hungarians, Ukrainians and Poles.41 The average frequency of R1a in Croatian mainland population (34%) is similar to the value of other Slavs, for example, Macedonians (35%), Czechs and Slovaks (27%).41 Figure 2 presents a phylogenetic reconstruction of haplogroup R1a haplotypes occurring three or more times. The most frequent one (16–12–10–27–25–10–11–13) has been observed in all populations except in Hvar. Among all R1a haplotypes samples from the northern island of Krk and mainland Croatia share the highest number of haplotypes, whereas the southern island of Hvar shows the lowest number of shared haplotypes.
SNP-based branching pattern of clades I, G and unresolved derivatives of haplogroup F did not recapitulate in MJ using eight STRs (data not shown). Three haplotypes were shared between haplogroups I and G, and a number of M170 haplotypes were reconstructed as descending from M201 haplotypes, in addition, M89* haplotypes were dispersed over the network lacking apparent phylogenetic structure.
Haplogroups G, E and J have been associated with the contribution of Neolithic farmers to the European gene pool.41,45 Decreasing frequencies of haplogroup G from the Middle-East to Europe have been observed with the highest frequency noticed in Georgia. Its frequency in the investigated Croatian population is low with the exception of the most southern and the most distant investigated island Korčula (10%), suggesting that the Neolithic spread may have come by sea rather than by land. Haplogroup J was found at low frequencies (0–4%) over all Croatia except of Krk where 11% of the lineages had an insertion at 12f2 locus. This low frequency of J is similar to Croatian northern neighbors Slovenians,33 but it is sharply in contrast to high frequencies of the cluster, often associated with M172,45 in other Mediterranean populations where >20% frequencies are common.33,41,46 Haplogroup E was found in Croatian populations in generally low frequencies as well, ranging from 4% in the south to 7% in Krk. This frequency is even lower than reported for other south European populations (Calabrian 16.2%, Sardinian 10.4%, Greek 24%, Macedonian 15%), but comparable or even higher than its 2–9% frequency in Czechs, Hungarians, Poles, Germans, northern Italians and Ukrainians.41
Discussion
The southern eastern Adriatic islands of Brač, Hvar and Korčula (Figure 1) had the highest frequencies reported in Europe to date (54–66%) of haplogroup I defined by M170. According to Underhill et al45 this mutation originated in Europe before the Last Glacial Maximum (LGM) and might have spread together with the Gravettian culture. Although, the frequency of its ancestral F* lineages is very low in Europe,45 their traces have been found in the investigated Croatian population (1.3%). During the LGM the Balkans,47 probably along with the northern Adriatic plain then connected with the present-day islands, could have been among a few refugia of Europe (eg Franco-Cantabria and Ukraine). High frequencies of mutation M170 noticed in the Croatian population investigated in this study and Bosnian and Herzegovian population (our unpublished data) imply that the Western Balkan Peninsula could have been an LGM reservoir of M170, a starting point of an expansion that spread M170 around the neighboring populations. Postglacial re-expansion from a different refugia might explain the differences of the northern European STR founder types from those characteristic to Croats.
The second most frequent haplogroup in mainland and island populations was R1a (frequency ranging from 9 to 38%). This haplogroup is almost completely absent in western Europe but widely present in eastern Europe and central Asia.33,41,44 The same geographic pattern can be seen in Croatia. A striking north–south difference implies that at least some of the founding ancestral groups of the investigated islands are of different origin. Possibly, continental Croatia was inhabited by Indo-European speaking populations who had migrated from southern Russia 2000 BP48,49,50 and probably had carried this mutation. It is logical that Dinarides are a natural mountain barrier to migrations between the southern Croatia and north-eastern Europe and possibly explaining the lower frequency of R1a lineages in the southern Adriatic islands of Brač, Hvar and Korčula.
Haplogroups R1b and I are likely to have been present in Europe since the Palaeolithic age.41 The frequency of R1b in Europe shows a major west-to-east decline being most frequent in the Basque. The observed frequency of R1b in Croatian sample (7.4%) harmonizes with its frequency in neighboring populations, although general frequency pattern runs also through the northwest to southeast directional layout of the Croatian populations showing a marked difference between mainland and northern Adriatic island of Krk and three southern Adriatic island groups (Brač, Hvar and Korčula). This indicates that migrations carrying M173 mutation were more pronounced in the northern part of Croatia than in the southern coastal area.
Worthy of note is the finding of considerable frequency of haplogroup P* (xM173) in the population of the island of Hvar. According to Wells et al44 this lineage displays a maximum in Central Asia while being rare in Europe, Middle East and East Asia. Its presence in Hvar recapitulates our finding of mtDNA haplogroup F on the island of Hvar and in mainland Croatian population that is virtually absent in Europe but, again, common in populations from Central and Eastern Asia.51 There are several possibilities for the occurrence of the ancestral lineage of M173. One is the well-documented alliance of Avars (a Mongol people) and Slavs (Croatians) that followed Avar arrival to the eastern Adriatic in 6th century AD. The other is the expansion of the Ottoman Empire from the 16th to 18th century AD when refugees from the western Balkan frequently immigrated to the islands. Lastly, the ancient Silk Road linking China with western Asia and Europe could be a possible path of P(xM173) lineage, too. Any of these migratory patterns could have introduced this mutation to the investigated population.
The observed frequency of the haplogroups E, G and J in Croatian sample is low, suggesting a minor genetic impact from Middle East. The minor Neolithic input in Croatia from the Near East as seen in frequency distribution of Y chromosome haplogroups is in concordance with the archaeological findings in this region. Chapman and Müller52 perceive the absence of late Mesolithic Q1sites in the area of the spread of early Neolithic sites in Greece and southern Italy; however, late Mesolithic sites do occur in Eastern Adriatic coast (known as Dalmatia) and further up in north and east parts of Croatia. Also, King and Underhill53 noticed in the Croatian region absence of distribution of Neolithic figurines and painted pottery that are associated with early agriculture period. This finding could imply that in this region transition to agriculture occurs later and less because of a replacement than diffusion. Highest frequency of haplogroup G in Korčula could be either because of drift or reflect a different source of migration, likely over the sea. A markedly higher frequency of haplogroup J noted at the northern island Krk might be a consequence of the fact that the island was most exposed to the immigrations from the mainland. During the Ottoman invasions to the Balkans population groups from the Adriatic hinterland (Dinarides) and Western Balkan area sought refuge at the eastern Adriatic islands. These populations often referred to as ‘Vlachs’ or ‘Morlachs’ included a few families of Romanian origin (sometimes described as descendants of the Roman army veterans as well) who left numerous traces of their presence at Krk (eg Skok54).
The lack of haplogroup N and its Tat C daughter clade in Croatians suggests that proto-Slavic males probably did not carry this lineage in substantial frequency and that Russians and Poles have likely obtained it through a recent admixture with Finno-Ugric people.34,55
In conclusion, the investigated Croatian populations show the presence of Y chromosomal haplogroups specific to Western, Southern and Eastern Europe. Moreover, Croatian Y chromosomal lineages testify to different migrational movements carrying mostly Palaeolithic European ancestry, a minor Neolithic impact from the Near East, as well as a Slavic (Croatian) influence which is today clearly expressed in the Croatian language which belongs to the Southern Slav linguistic group. Haplogroup I, one of the few haplogroups of Palaeolithic European origin, present in Croatians in the highest frequency noticed in Europe so far, could potentially classify this area as a birth place of this mutation as well as a source of its post-LGM spread in Europe. Our data (12.5%) do not support the estimation of more than 82% of Neolithic contribution in Croatian population suggested by Chikhi et al.56 Chikhi et al56 assume homogeneity of Palaeolithic gene pool in Europe by taking only the Basques as representatives of Palaeolithic Europeans thus defining all other lineages by default as Neolithic. This assumption is likely not to hold, at least for haplogroup I, because it is restricted to Europe and almost absent in the Middle-East.
Having in mind a gradual geographic appearance of haplogroups R1a and R1b and domination of I in southern Adriatic islands of Brač, Hvar and Korčula one could expect that the ancestral founding groups of investigated populations could be of different origins. The genetic legacy of Adriatic islands corresponds to the proximal mainland and not to the neighboring Mediterranean populations, although the archaeological evidence suggests a different pattern. Apparently, observed genetic background of the studied southern islands area might be primarily structured by a population dispersal directed from the mainland towards the islands, whereas a cultural exchange was transferred mainly via Mediterranean maritime routes.
A general conclusion of previous anthropological studies was that the islands more distant from the coast (Hvar and Korčula) were the ones exhibiting higher degree of isolation, endogamy and inbreeding.13,57 This study showed that Y chromosome haplotype diversity on Korčula island was lower than in other islands (0.9749±0.0059 in comparison with the northern island Krk 0.9882±0.0047, and southern islands of Brač 0.9787±0.0099 and Hvar 0.09839±0.0049). This is consistent not only with the findings based on endogamy and inbreeding, but also with the implications of our previous mtDNA studies,16 stressing the influence of drift in small isolated Adriatic island populations.
Finally, clear and meaningful in terms of ethnogenesis pattern and gradients of Y chromosome distribution variants in Croatia, uniparentally inherited genetic loci do not only prove their usefulness in understanding demographic history of human populations, but also indicate the need for their evaluation within the context of isonymic analyses.
References
Jobling MA, Tyler-Smith C : Fathers and sons: the Y chromosome and human evolution. Trends Genet 1995; 11: 449–456.
Hammer MF, Zegura SL : The role of the Y chromosome in human evolutionary studies. Evol Anthropol 1996; 5: 116–134.
Underhill PA, Jin L, Lin AA et al: Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res 1997; 7: 996–1005.
Underhill PA, Shen P, Lin AA et al: Y chromosome sequence variation and the history of human populations. Nat Genet 2000; 26: 358–361.
Clarke AG : Natural selection and Y-linked polymorphism. Genetics 1987; 115: 569–577.
Kimura M : The neutral theory of molecular evolution. Cambridge: Cambridge University Press, 1983, pp 40–50.
Rudan P : Microevolution studies in present populations in Yugoslavia. Coll Antropol 1980; 4: 35–39.
Rudan P, Roberts DF, Janićijević B, Smolej N, Szirovicza L, Kaštelan A : Anthropometry and the biological structure of the Hvar population. Am J Phys Anthropol 1986; 70: 231–240.
Rudan P, Angel JL, Bennett LA : Historical processes and biological structure of the populations – example from the island of Korčula. Acta Morphol Nether Scand 1987; 25: 69–82.
Rudan P, Šimić D, Smolej-NaranČić N et al: Isolation by distance in Middle Dalmatia – Croatia. Am J Phys Anthropol 1987; 74: 417–426.
Rudan P, Šimić D, Bennett LA : Isolation by distance on the island of Korčula: correlation analysis of distance measures. Am J Phys Anthropol 1988; 77: 97–103.
Rudan P, Sujoldžić A, Šimić D, Bennett LA, Roberts DF : Population structure in the eastern Adriatic: the influence of historical processes, migration patterns, isolation and ecological pressures, and their interaction; in Roberts DF, Fujiki N, Torizuka K (eds): Isolation, migration and health. Cambridge: Cambridge University Press, 1992, pp 204–219.
Rudan P, Janićijević B, Šimić D, Bennett LA : Migrations in the Eastern Adriatic and their consequences on population structure – analyses of census data; in Lopašić A (ed): Mediterranean societes: transition and change. Zagreb: HAD, 1994, pp 113–122.
Š imić D, Rudan P : Isolation by distance and correlation analysis of distance measures in the study of population structure: example from the island of Hvar. Hum Biol 1990; 62: 113–130.
Waddle DM, Sokal RR, Rudan P : Factors affecting population variation in Eastern Adriatic isolates (Croatia). Hum Biol 1998; 70: 845–864.
Tolk H-V, Peričić M, Barać L et al: MtDNA Haplogroups in the populations of Croatian Adriatic islands. Coll Antropol 2000; 24: 267–279.
Malez M : Palaeolithic Q2and Mezolithic regions and cultrues in Croatia; in Benac A (ed): Prehistory of south Slavic countries. Sarajevo: Svjetlost, 1979, 1979, vol 1, pp 277–295.
Š egota T : Paleogeographic changes in the Adriatic sea from Würm Glacial Maximum to recent (in Croatian). Radovi 1982–1987; 1118: 11–15.
Forenbacher S : Prehistoric populations of the island of Hvar – An overview of archeological evidence. Coll Antropol 2002; 1: 361–378.
Zilaho J : From the Mesolithic to the Neolithic in the Iberian peninsula; in Price TD (ed): Eruope's first farmers. Cambridge: Cambridge University Press, 2000, pp 144–182.
Müller J : Das Ostadriatische Frühneolitikum: Die Impresso-Kultur und die Neolithiserung des Adriaraumes. Berlin: Volker Spiess, 1994.
Poncz M, Solowiejczyk D, Harpel B, Mory Y, Schwartz E, Surrey S : Construction of human gene libraries from small amounts of peripheral blood: analysis of beta-like globin genes. Hemoglobin 1982; 6: 27–36.
Hurles ME, Irwen C, Nicholson J et al: European Y-chromosomal lineages in Polynesians: a contrast to the population structure revealed by mtDNA. Am J Hum Genet 1998; 63: 1793–1806.
Hammer MF : A recent insertion of an alu element on the Y chromosome is a useful marker for human population studies. Mol Biol Evol 1994; 11: 749–761.
Hammer MF, Horai S : Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet 1995; 56: 951–962.
Whitfield LS, Sulston JE, Goodfellow PN : Sequence variation of the human Y chromosome. Nature 1995; 378: 379–380.
Kwok C, Tyler-Smith C, Medonca BB et al: Mutation analysis of 2 kb 5′ to SRY in XY females and XX intersex subjects. J Med Genet 1996; 33: 465–468.
Hammer MF, Karafet T, Rasanayagam A et al: Out of Africa and back again: nested cladistic analysis of human Y chromosome variation. Mol Biol Evol 1998; 145: 427–441.
Mathias N, Bayes M, Tylor-Smith C : Highly informative compound haplotypes for the human Y chromosome. Hum Mol Genet 1994; 3: 115–123.
Hurles ME, Veitia R, Arroyo E et al: Recent male-mediated gene flow over a linguistic barrier in Iberia, suggested by analysis of a Y-chromosomal DNA polymorphism. Am J Hum Genet 1999; 65: 1437–1448.
Santos FR, Pandya A, Tyler-Smith C et al: The central Siberian origin for native American Y chromosomes. Am J Hum Genet 1999; 64: 619–628.
Casanova M, Leroy P, Boucekkine C et al: A human Y-linked DNA polymorphism and its potential for estimating genetic and evolutionary distance. Science 1985; 230: 1403–1406.
Rosser Z, Zerjal T, Hurles ME et al: Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 2000; 67: 1526–1543.
Zerjal T, Dashnyam B, Pandya A et al: Genetic relationships of Asians and northern Europeans, revealed by Y-chromosomal DNA analysis. Am J Hum Genet 1997; 60: 1174–1183.
The Y Chromosome Consortium: A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 2002; 12: 339–348.
Cruciani F, Santolamazza P, Shen P et al: A back migration from Asia to Sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet 2002; 70: 1197–1214.
deKnijf P, Kayser M, Caglia A et al: Chromosome Y microsatellites: population genetic and evolutionary aspects. Int J Leg Med 1997; 110: 134–140.
Schneider S, Kueffer J-M, Roessli D, Excoffier L . Arlequin version 1.1: a software for population genetic data analysis. Switzerland: Genetics and Biometry Laboratory, University of Geneva, 1997.
Richards M, Macaulay V, Hickey E et al: Tracing European founder lineages in the near eastern mtDNA pool. Am J Hum Genet 2000; 67: 1251–1276.
Bandelt HJ, Forster P, Rohl A : Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 1999; 6: 37–48.
Semino O, Passarino G, Oefner PJ et al: The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 2000; 290: 1155–1159.
Passarino G, Cavalleri GL, Cavalli-Sforza LL et al: Different genetic components in the Norwegian population revealed by the analysis of mtDNA and Y chromosome polymorphisms. Eur J Hum Genet 2002; 10: 521–529.
Helgason A, Sigurethardottir S, Nicholson J et al: Estimating Scandinavian and Gaelic ancestry in the male settlers of Iceland. Am J Hum Genet 2000; 67: 697–717.
Wells RS, Yuldasheva N, Ruzibakiev R et al: The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA 2001; 98: 10244–10249.
Underhill PA, Passarino G, Lin AA et al: The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 2001; 65: 43–62.
Scozzari R, Cruciani F, Pangrazio A et al: Human Y-chromosome variation in the western Mediterranean area: implications for the peopling of the region. Hum Immunol 2001; 62: 871–884.
Hewit G : The genetic legacy of the Quaternary ice ages. Nature 2000; 405: 907–913.
Jovanović B : Steppe culture in Eneolithic period Q2in Yugoslavia; in Tasić N (ed): Prehistory of south Slavic countries. Sarajevo: Svjetlost, 1979, vol 3, pp 381–396 (in Croatian).
Jovanović B : Indo-Europeans and Eneolithic period in Yugoslavia; in Tasić N (ed): Prehistory of south Slavic Q2countries. Sarajevo: Svjetlost, 1979, vol 3, pp 397–416 (in Croatian).
Tasić N, Dimitrijević S, Jovanović B : Final conclusions; in Tasić N (ed): Prehistory of south Slavic countries. Sarajevo: Svjetlost, 1979, vol 3, pp 417–460 (in Croatian).
Tolk H-V, Barać L, Peričić M et al: The evidence of mtDNA haplogroup F in a European population and its ethnohistoric implications. Eur J Hum Genet 2001; 9: 717–723.
Chapman J, Müller J : Early farming in the Mediterranean Basin: the Dalmatian evidence. Antiquity 1990; 64: 127–134.
King R, Underhill PA : Congruent distribution of Neolithic painted pottery and ceramic figurines with Y-chromosome lineages. Antiquity 2002; 76: 707–714.
Skok P : Slavenic and Romanic influence on the Adriatic islands. JAZU, Zagreb: Jadranski institut, 1950 (in Croatian).
Laitinen V, Lahermo P, Sistonen P, Savontaus ML : Y-chromosomal diversity suggests that Baltic males share common Finno-Ugric-speaking forefathers. Hum Hered 2002; 53: 68–78.
Chikhi L, Nichols RA, Barbujani G, Beaumont MA : Y genetic data support the Neolithic demic diffusion model. Proc Natl Acad Sci USA 2002; 99: 11008–11013.
Rudan I, Campbell H, Rudan P : Genetic epidemiological studies of Eastern Adriatic island isolates, Croatia: objectives and strategies. Coll Antropol 1999; 23: 531–546.
Acknowledgements
We thank all the subjects for their cooperation in performing this study. We thank Jaan Lind for technical assistance. This work was supported by the Ministry of Science and Technology of the Republic of Croatia grant to P Rudan (project title: Population structure of Croatia – anthropogenetical approach, No. 0196005), EC ICA-CT-2000-70006 grant to R Villems and field research by Wellcome Trust to I Rudan.
Author information
Authors and Affiliations
Corresponding author
Additional information
The first two authors have equally contributed to this work.
Rights and permissions
About this article
Cite this article
Barać, L., Peričić, M., Klarić, I. et al. Y chromosomal heritage of Croatian population and its island isolates. Eur J Hum Genet 11, 535–542 (2003). https://doi.org/10.1038/sj.ejhg.5200992
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200992
Keywords
This article is cited by
-
Croatian national reference Y-STR haplotype database
Molecular Biology Reports (2012)
-
Distinguishing the co-ancestries of haplogroup G Y-chromosomes in the populations of Europe and the Caucasus
European Journal of Human Genetics (2012)
-
Y-chromosome distributions among populations in Northwest China identify significant contribution from Central Asian pastoralists and lesser influence of western Eurasians
Journal of Human Genetics (2010)
-
3000 years of solitude: extreme differentiation in the island isolates of Dalmatia, Croatia
European Journal of Human Genetics (2006)
-
Y-Haplotype X in the Balkans
International Journal of Anthropology (2006)