Abstract
A thorough study of the heterochromatin organisation in the pericentromeric region and the proximal long (q) and short (p) arms of human chromsome 9 (HSA 9) revealed homology between 9p12 and 9q13-21.1, two regions that are usually not distinguishable by molecular cytogenetic techniques. Furthermore, the chromosomal regions 9p12 and 9q13-21.1 showed some level of homology with the short arms of the human acrocentric chromosomes. We studied five normal controls and 51 clinical cases: 48 with chromosome 9 heteromorphisms, one with an exceptionally large inversion and two with an additional derivative chromosome 9. Using fluorescence in situ hybridisation (FISH) with three differentially labelled chromosome 9-specific probes we were able to distinguish 12 heteromorphic patterns in addition to the most frequent pattern (defined as normal). In addition, we studied one inversion 9 case with the recently described multicolour banding (MCB) technique. Our results, and previously published findings, suggest several hotspots for recombination in the pericentromeric heterochromatin of HSA 9. They also demonstrate that constitutional inversions affecting the pericentromeric region of chromosome 9 carry breakpoints located preferentially in 9p12 or 9q13-21.1 and less frequently in 9q12.
Similar content being viewed by others
Introduction
Among the non-acrocentric human chromosomes, chromosome 9 presents with the highest degree of morphological variations.1 Variants, such as 9qh+, 9qh− or inv(9)(p11q13) are very common findings in routine cytogenetics, with frequencies of approximately 8%, <1% and approximately 1.5%, respectively.2 These variants, termed ‘heteromorphisms’, have been studied intensely and are usually ascertained by GTG- and CGB-banding in routine cytogenetics.2 The ISCN 19953 describes the following variants for chromosome 9: (i) increase or decrease in the length of the heterochromatin of the long arm (9qh+ and 9qh− respectively), and (ii) heterochromatin in the short arm only (9ph), or in both the long and the short arm (9phqh). Surprisingly, the most common inversion, inv(9)(p11q13), cytogenetically classified as a variable but normal chromosome feature4 is not mentioned in the ISCN 1995.3
Since the introduction of fluorescence in situ hybridisation (FISH) techniques only a few systematic studies have been performed to classify the chromosome 9 heteromorphisms in more detail. In one study, four different types of inversion and duplication (ie 9qh+) were reported,5 and overall, seven different types of rearrangement leading to inversions on chromosome 9 have been described.1,6,7 However, neither the origin nor the clinical significance of these heteromorphisms is well understood. This is mainly due to the fact that the centromeric regions of the human chromosomes are poorly assembled by current mapping and sequencing efforts, as they are poor in euchromatin but repeat-rich and organised in a highly complex fashion.8
The present study used a specific probe set of three chromosome 9-specific FISH probes (chromosome 9 α-satellite, satellite III DNA and a microdissection-derived probe for 9q12/9q13-21.1) to identify the full range of heteromorphic patterns and any breakpoint cluster regions related to the pericentric inversions of chromosome 9. We also wanted to assess whether the multicolour banding (MCB) technique would detect any heteromorphic patterns of clinical significance.
Patients and methods
Patients
The study group was chosen based on previous findings of heteromorphic patterns in the pericentric region of chromosome 9. Informed consent for the study was obtained from all patients. Metaphase spreads from short-term cultures of peripheral blood or aminocytes from a total of 56 patients were analysed by standard GTG- and CGB-banding in eight German Centres (Cottbus, Düsseldorf, Essen, Halle/Saale, Homburg/Saar, Jena, Leipzig and Magdeburg; Table 1). With the exception of case 52, who was the mother of case 56, patients were not related. A familial 9ph+ heteromorphism (defined by GTG-banding) was found in metaphases from cases 52 and 56, but the child also carried a supernumerary marker chromosome (SMC) of chromosome 9 origin. Case 12 was the partner of case 16.
Twenty-five healthy patients were analysed cytogenetically for reasons not relevant in the context of our study. Another 17 patients were analysed due to a history of reproductive problems. One patient was healthy but cytogenetic analysis revealed an SMC in 75% of the metaphases (case 55). In 12 cases, the chromosome 9 variants were detected in children or young adults in connection with minor physical abnormalities or psychomotor retardation (defined as morbid slowing of action or movement presumed to result from psychic rather than physical disturbance). Although it could not be verified, it is probable that the heteromorphisms detected in these cases were familial variants.
Molecular cytogenetics
The chromosomes of all 56 patients were studied using a FISH-probe set comprised of: (a) a chromosome 9-specific a-satellite probe (SpectrumGreen, green; Vysis, Downers Grove, IL, USA); (b) a satellite III probe (digoxigenin, detected with anti-digoxigenin-rhodamine, red; Q-BIOgene, Heidelberg, Germany); (c) a microdissection-derived9 probe from the SMC of case 56 (‘midi36’), specific for 9p12 and pq13-21.1; diethylaminocoumarin, blue (NEN-Dupont, Boston, MA, USA). Additionally, a microdissection derived whole chromosome and chromosome arm-specific painting probes,10 microdissection probe for all human acrocentric chromosomes (‘midi’54) (derived from the SMC of an ICSI patient and representing heterochromatin from the short arms of chromosome 13 or 21),11 and a probe specific for all human centromeres (Q-BIOgene) have been applied in some of the cases. Labelling of the microdissection derived probes and hybridisation were carried out as described.10,12 We used a chromosome 9-specific MCB probe set11 to further characterise the heteromorphic chromosome 9 in case 13 following the published MCB FISH procedure.11,13 All images were captured on a Zeiss Axioplan microscope (Zeiss, Jena, Germany) with the IKAROS and ISIS digital imaging system (MetaSystems, Altlussheim, Germany) using a CCD camera with on-chip integration (XC77, Sony, Vienna, Austria).
Results
A chromosome 9-derived small marker chromosome
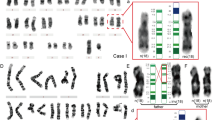
The DNA-probe midi36 (derived from the SMC of case 56) stained the chromosomal regions 9p12 and 9q13-21.1 on both normal chromosomes 9 in addition to the SMC (Figures 1A and 2C). Both the mother (case 52) and child (case 56) carried one chromosome defined cytogenetically at 9ph+ (Figures 1A and 2B caused by a pericentric inversion with breakpoints in 9p12 and 9q12−13 (Figures 1A2. In addition to the SMC, the child carried one normal chromosome 9 and one chromosome with a ‘9ph+’ heteromorphism. It is possible that the SMC originated from the region composed of 9p12 and 9q13-21.1 (Figure 1A2). However, no further studies of the origin of the SMC could be performed due to the unavailability of a parental sample. Hybridisation of probe midi36 and post-hybridisation slide processing under less stringent conditions results in signals in the pericentromeric regions of most human chromosomes including 1qh and 16qh, and the short arms of the acrocentric chromosomes (Figure 1E). Cohybridisation of midi36 with a microdissection-derived probe for the p-arms of all acrocentric chromosomes (midi54)11 highlighted identical regions on chromosome 9 (Figure 1F). The probe midi54 gave the signals only on the pericentromeric region of chromosome 9 regardless of stringency levels (Figure 1F).
Representative examples of the FISH patterns observed. (A) (1) Representative partial metaphase from the foetus (case 56) carrying the SMC from which the microdissection probe midi36 was derived. In addition to painting the SMC itself (arrowhead), this probe allowed the identification of the heteromorphic pattern (var2) (9qh+ and inversion) for the chromosome 9 cytogenetically defined as ‘9ph+’. (2) Schematic depiction of the postulated origin of the SMC of case 56. A pericentric inversion with breaks in distal 9p12 and proximal 9q13 led to the ‘9ph+’ variant detected in case 52 (mother of case 56). The SMC is a derivative of that variant of chromosome 9 with breaks in 9p10 and 9q13-21.1 (B) Partial metaphase from case 6 of the probe midi36 (red) co-hybridised with a classical satellite III DNA probe (green). The reduced green signal on the ‘qh-’chromosome indicates that this is due to a diminished copy number of the classical satellite III DNA. (C) Representative chromosomes from case 14 hybridised with: (1) a whole chromosome 9 paint (wcp 9) (green) and a classical satellite III DNA probe (red). The CBG negative GTG-positive extra-band in 9q12 is highlighted by wcp 9, and (2) the midi36 (green) probe. (D) Cohybridisation of the chromosome 9 specific α-satellite DNA probe used in the present study (red) with an alpha satellite probe (pan α-sat, green) specific for all human centromeres. The chromosome 9 specific α-satellite DNA probe covers a similar but larger region to the pan α-satellite probe. (E) Under low stringency conditions the midi36 probe hybridized to homologous sequences in the pericentric region of most human chromosomes including 1qh and 16qh, and the p-arms of the acrocentric chromosomes. Under the same low stringency hybridisation conditions, the satellite III DNA probe (green) highlighted only the two chromosomes 9. (F) (1) Metaphase showing cohybridisation of the midi36 probe (red) and a microdissection probe for the acrocentric p-arms (midi54, green). The enlarged chromosomes in (2) show identifcal regions of chromosome 9 labelled by both probes. (G) The var3 (9qh+ and inversion) type inversion of case 14 analysed using the multicolor banding (MCB) probe set for chromosome 9. Using the MCB probe set, the α-satellite region is pseudo-colored in pink, the satellite III DNA region in white and the region 9p12/9q13-21.1 in violet. These results confirm those obtained with a set of three centromeric chromosome 9 probes (Figure 2).
Representative FISH patterns and schematic representative of heteromorphic patterns observed in the study. (A) The 12 observed heteromorphic patterns and a ‘normal’ variant detected by a probe set of chromosome 9 specific alpha satellite DNA (green), classical satellite III DNA (red) and midi36 (blue) are shown. On the left hand side, the Legend shows a schematic version of the normal, qh+, qh-, ph+, and five variant types of inversion (inv var1–5). The number of cases detected with the corresponding heteromorphic pattern is given below each group. The suggested type of rearrangement leading to the 12 heteromorphic pattern is given in grey near the arrows. (B) Special hybridization patterns observed in the ring chromosome of case 55 and the derivative chromosome 9(p10q12) of case 56. The hybridisation pattern of the large inv(9)(p24.1q31.2) of case 54 (big inv) using the probe set of (A) (left) and partial chromosome painting probes specific for 9p (green) and 9q (red) is also shown. (C) Localisation of the probes used in the present study and in former studies. (D) Summary of the heteromorphic pattern detected in the studies of Ramesh and Verma6 and Samonte et al.7 Abbreviations: inv=inversion; add of sat III=addition of material; del=deletion of material; sat III=classical satellite III DNA; pan-α-sat= pan-alpha-satellite DNA probe; α-sat 9=chromosome 9 specific alpha-satellite DNA probe; β-sat=chromosome 9 specific beta satellite DNA probe; sat III=satellite III DNA probe.
The probe midi36 completely stained the CBG-negative/GTG-positive extra band in 9q12 in cases 13–17 (Figure 1C). Combining midi36 together with a satellite III DNA probe showed that the size variations 9qh+ and 9qh− are due to variations in copy number of the satellite III DNA (Figures 1B and 2A).
Characterisation of heteromorphic patterns
In cases characterised as 9qh+, 9ph+, or inv(9)(p11q13), 12 different heteromorphic patterns (besides the normal) could be distinguished using our three probe set. The results are summarised in Figures 2A–C, 3 and Table 1.
Mechanisms of formation of heteromorphisms observed. Six theoretically possible breakpoints in the two regions stained by the probe midi36 are postulated: a proximal (black arrowhead), centric (dark-grey arrowhead) and distal breakpoint (light-grey arrowhead) in 9p12, and a proximal (light-grey arrow), centric (dark-grey arrow) and distal breakpoint (black arrow) in 9q13-21.1. Therefore, nine different breakpoint-combinations are possible (A–E). Additionally, breakpoints in 9q12 (green arrowhead) and in the alpha-satellite region (blue arrowhead) are postulated (see I). In F–H the postulated breakpoints for the development of the inversion types var3, var4 and var5 are given (for further details refer to the text). (Abbreviations: bp=breakpoints; nd, nd1, nd2=different possible variants which have not been detected in the present study).
Previous studies6,7 of the 9qh region used a satellite III DNA probe combined with a β-satellite probe and an α-satellite probe specific for all human centromeres (pan-α satellite probe; Figure 2C,D). In the present study, we compared the patterns obtained with the chromosome 9 specific α satellite probe with the pan-α satellite probe by cohybridisation. As shown in Figure 3, the chromosome 9 specific α satellite probe covers a similar but larger region to the pan-α satellite probe (Figure 1D).
The normal pattern
All 10 homologues of chromosomes 9 in the control group (cases 1–5) and one chromosome each of the 51 patients studied showed the following hybridisation pattern: two blue signals (corresponding to the midi36 probe) in 9p12 and 9q13−21.1 respectively and flanking (from p to q) (i) a small green α satellite and (ii) a red satellite III signal. If the latter was of about the same size as the sum of both midi36 signals (for the normal pattern see Figures 2A and 3A–C, upper row), the region 9q12 was considered normal. The reqion 9q12 was defined as 9qh− if the red band was smaller than either blue midi36 signals, and as 9qh+ if the red band was larger than the sum of midi36 signals.
Abnormal patterns: inv(9)(p11q13) and 9qh−
Thirty-two cases (cases 18–49) classified as inv(9)(p11q13) by GTC- and CBG-banding could be sub-classified based on three different FISH patterns (Figure 2A). All 32 cases showed an inverted pattern of FISH signals. Twelve cases (cases 18–29) presented with blue signals of equal size at 9p12 and 9q13-21.1 (variation type 1=var1). Twenty cases showed a weaker signal at 9p12 and a large signal at 9q13-21.1 (variation type 2=var2, cases 30–49). These two groups could be divided further into: (a) cases with a satellite III region of normal size and (b) cases with a red fluorescent signal of reduced size (less than half the size of the midi36 signals). This was defined as 9qh− with inversion. Only one case of this variant was found in the patients studied. While var1 was present in only six of the 26 cases with inversion and a satellite III region of normal size, var1 was found in all six 9qh− cases with inversion (Table 1).
The 9ph+ pattern
Five cases (cases 50–53, 56) were described as 9ph+ by GTG banding. Cases 50 and 51 showed a duplication of either the region 9p12 or 9q13-21.1, which are indistinguishable in our model. In case 53, a similar enlargement was followed by an inversion with breaks in 9q13-21.1 and in 9q12 (inversion type var3). In case 52 and her unborn child (case 56) FISH revealed an enlargement of satellite III DNA in connection with a pericentric inversion of var2 type as cause of the 9ph+ (see Figures 1A and 2).
The 9qh+ pattern
We included eleven cases described as 9qh+ after cytogenetic analysis (cases 7–17). Five of these had an enlarged satellite III (red) signal but no aberrant pattern for the other two probes. In all 9qh+ cases, the enlarged heterochromatin in the pericentric region of chromosome 9 was larger than the sum of blue midi36 signals. In five cases an extra band was observed on GTG-banding (cases 13–17; Table 1). This additional band was delineated by the probe midi36 (see Figures 1C and 2A). Three of these (cases 13–15) showed a normally sized blue signal in 9p12, and a proximal green signal, but two blue signals in the long arm of chromosome 9 (one in 9q12 and 9q13-21.1, respectively). The signal in 9q13-21.1 was less intense than that in 9p12, but both signals in 9q together were as intense as that in 9p12. This pattern could be explained by a paracentric inversion with breaks in 9q12 and 9q13 (inversion variation 3=var3).
In case 16 two additional signals for the probe midi36 were present in the long arm of chromosome 9. This pattern can be explained by the occurrence of a second inversion (after the var3 inversion), with breakpoints in proximal 9q12 and the midi36 positive extra band (inversion type 4=var4) (see Figure 2A). The hybridisation patterns found in case 17 are also most likely due to a second inversion following a var3 inversion. In this case the breakpoints are likely to be in 9q11 and the midi36 positive extra band on the var3, leading to split signals within the α satellite region (inversion type 5=inv5) (see Figure 2A).
Additional aberrant pattern
There were three cases of abnormalities detected in apparently healthy individuals. The first (case 54), was discovered to have an exceptionally large pericentric inversion with breakpoints in 9p24.1 and 9q31.2 (Figure 2B). Although the true constitution of this rearrangement could not be elucidated using our probe set, the observed pattern was similar to an inversion type var1 (Figure 2B). The second, an SMC detected in a healthy women (case 55), was characterised as a ring chromosome comprised exclusively of α-satellite and satellite III DNA (ring q12) (see Figure 2B). The third was the SMC of case 56, described above.
A recently developed multicolour banding approach11,13 using a chromosome 9 specific probe set11 was applied to analyse the inversion type 3 in case 14 (Figure 1G). The MCB results confirmed results obtained with the set of three chromosome 9 DNA repeat probes (Figure 2A).
Mechanism of formation of 9q heteromorphisms
Six potential breakpoints are postulated within the two regions stained by the probe midi36: a proximal, a centric and a distal breakpoint in 9p12 and analogous regions in 9q13-21.1. Therefore, nine different breakpoint combinations are possible, three of which could result in the ‘var1’ and two in the ‘var2’ heteromorphic pattern (Figure 3A,B respectively). Only one breakpoint combination could result in a pattern similar to that observed in the 9ph+ cases (Figure 3C). It is possible that the enlarged signal in 9q13-21.1 corresponding to the midi36 probe in the 9ph+ cases 50, 51 and 53 is an artifact due to flaring effects of the microdissection-derived probe, previously described by Speicher et al. for whole chromosome painting probes.14 However, due to the consistency of the signal size we believe that this reflects amplification of 9p12 in 9ph+ karyotypes. Three of the possible breakpoint combinations lead to signal patterns not observed in the present study (Figure 3D,E). It remains to be determined if these variants exist or if there is a specific mechanism that prohibits their formation. Additionally, a combination of the postulated centric breakpoint in 9q13-21.1 and a breakpoint in 9q12 would lead to the inversion type ‘var3’. This was present in four cases: three of which were 9qh+ (cases 13–15) and one a 9ph+ (case 53). The inversion types 9qh+/var4 and 9qh+/var5 can easily be traced from 9qh+/var3 by postulating breaks in the proximal region of 9q13-21.1 and 9q12 or in 9q12 and 9q10, respectively (Figure 3F–H).
Discussion
Due to their paralogous organization, multiple gaps are still present in the primary sequence of the pericentromeric regions of chromosome 9.8 However, even without specific knowledge of the DNA sequence, molecular cytogenetic studies such as the one presented here can provide important information. The present study has confirmed the homology between 9p12 and 9q13-21.1 reported by other investigators15,16 (Figure 2B). The identification of 9qh+ individuals with an additional GTG-positive and CBG-negative band in the satellite III region at 9q12 (Figure 1C1) has also been reported previously.17,18 However, the observation that the midi36 (9p12) and midi54 (9q13-21.1) probes cross-hybridise with the short arms of the acrocentric chromosomes is a new finding, and suggests that these regions might be evolutionarily related. These observations confirm other evolutionary data concerning Homo sapiens (HSA) chromosome 9 homologues in primates.11 HSA 9 developed from an acrocentric ancestor by a pericentric inversion with breaks in regions homologous to 9p24.3 and 9q13.11 During further evolution a satellite III DNA block may have been inserted de novo into the DNA block stained by the probe midi36, and in some cases, it was subsequently amplified (9qh+) or diminished (9qh−) (Figure 4). The finding of homology between the 9p12/9q13-q21.1 regions and acrocentric short arms suggests that these regions might be more prone to chromosomal rearrangements. However, a search of the literature has shown that this is not the case, with only a single report of translocations involving these regions.19,20,21,22
Development of human chromosome 9 (HSA 9) from its presumed ancestor. HSA=homo sapiens. Schematic representation of the origin of HSA 9, based on hybridization of the midi54 probe to chromosomes of the Gorilla gorilla (GGO).11 The p-arm of acrocentric GGO 13 (homologue to HSA 9) has homologous sequences to the short arm of all human acrocentric chromosomes,11 the region stained by midi36 (blue). A pericentric inversion (inv) of GGO 13, with breaks in regions homologous to 9q24.3 and 9q13 leads to a human chromosome 9 banding pattern. During evolution human specific α-satellite DNA (red) has developed and inserted (ins) together with the satellite III DNA block (green).
This study has identified 12 heteromorphic patterns of the pericentromeric region of chromosome 9, in addition to the ‘normal’ variant (Figures 2A and 3). It is difficult to compare the heteromorphic patterns detected in the present study with other studies from the literature, due to the variety of techniques used, many of which lacked the resolution presented in this investigation. The inv(9)(p11q13) types var1 and var2 have not been described before. However, the unusual chromosome 9 polymorphism described by Fryns et al.23 in a healthy individual may be similar to the one found in our case 12 (9qh+ and inversion type var1). The heteromorphic patterns identified by Ramesh and Verma6 and Samonte et al.7 were detected with FISH using a pan-α-satellite and a chromosome 9 specific β-satellite DNA probe (see Figure 2D). However, as the β-satellite probe used in these studies is no longer available, we can only speculate that our inversions type var1 and var2 could be subdivided into the same subtypes. An acquired inv(9)(p11q13) has been described in essential thrombocytothemia,24 but the type of inversion is not evident from the literature.
Healthy individuals with a 9ph+ variant have been reported. In some of these (eg Family D in the study of Wang and Miller25), the additional material in 9p originates from the satellite III DNA, similar to case 52 in the present study. The cases described by Webb et al.26 show more similarity to cases 50, 51 and 53 of the present study. Interestingly, Webb et al.26 detected an amplification of the 9ph+ region transmitted through all generations. Reddy,27 Knight et al.28 and Jalal et al.29 all report healthy carriers with a CBG negative 9qh+ variant. This variant was not detected in the present study. The five cases of 9qh+ with simple amplification of satellite III DNA are similar to previously reported cases.30 Published cases of individuals with a GTG-positive/CBG-negative extra band in 9qh+27,31,32,33,34,35,36,37 probably correspond to 9qh+ and inversion of type (i) var3, (ii) var4 or (iii) var5. This study therefore indicates that the incidence of pericentric inversions involving the heterochromatin may be higher than the suggested 1.5%.38 Moreover, the results presented here indicate that the conclusions of Wojiski et al.36 and Park et al.15 that the additional material in the extra band in 9q12 is derived from 9p12 (based on the hybridisation pattern of YAC 878e3) are incorrect – it also could be derived from 9q13-21.1. Several variants have been described that do not fit any of the heteromorphic patterns described here. Verma et al.39 described an inverted duplication 9qh in a prenatal case. Inverted duplications of 9p10-q12 were also reported by Wang and Miller25 and Macera et al. (Type II)5 using FISH with a pan-α-satellite probe and a chromosome 9-specific β-satellite DNA probe. The inversion type C described by Samonte et al.7 is similar to our 9qh+ with inversion type var3, while their types A, B and D were not detected in our study.
A possible connection between the 9ph polymorphisms and reproductive problems has been postulated from the beginning of the banding era2,3,4 up to the present day.40,41 In our study, one couple (case 13 and 17) that underwent intra-cytoplasmic sperm injection (ICSI) for fertility problems carried a 9qh+ combined with inversion(s) (var3 and var5) in both partners. Furthermore, four of the five cases with 9qh+ and an extra band in 9q12 (including the couple mentioned before) reported fertility problems. Additionally, 17 out of the 47 cases studied with heteromorphism of chromosome 9 (36%) were referred for chromosome analyses because of reproductive failure or reduced fertility (ie repeated foetal miscarriages and/or ISCI patients). In a control group of 100 patients referred for cytogenetic analysis because of reproductive failure in Jena between December 2000 and January 2001, the corresponding percentage of cases without heteromorphism of chromosome 9 was 24%, providing further evidence for a possible correlation. Even more strikingly, 13 out of the 17 cases with heteromorphism of chromosome 9 referred because of reproductive failure were female (see Table 1). In contrast, the risk to male inv(9)(p11q13) carriers of producing chromosomally abnormal offspring or spontaneous abortions was not increased in the study by Colls et al.40 There are many and varied clinical conditions reported to have an association with pericentric inversion 9 including mental retardation,42 schizophrenia,43 the Walker-Warburg syndrome,44 the oculo-auriculo-vertebral (Goldenhar) spectrum45 and cancer predisposition.46 To date therefore, the clinical significance of the chromosome 9 heteromorphism still remains to be clarified.
The majority of the heteromorphic patterns revealed in this study (apart from 9qh+ or 9qh− copy number variations of classical satellite III DNA) can be explained by simple inversions with breakpoints in the repetitive elements of the regions 9p12 and 9q13-21.1 or in the satellite III region (Figure 3). The finding that the majority of these breakpoints are within repetitive sequence regions is important, and may explain why such inversions do not have any clinical consequences. There is also evidence to suggest hotspots for recombination in these chromosomal sections (see Figure 3). Large DNA sequence duplications flanking the centromere of human chromosome 10 and promoting pericentric inversions have been described recently.47 It is possible that sequence similarities of this type could form the molecular basis for inversions involving chromosome 9 as well. However, the large amount of repetitive DNA at the centromere of chromosome 9 has so far hampered mapping and sequencing efforts.8
In summary, 12 heteromorphic patterns and the normal variant of the pericentric region of chromosome 9 are described. Previous studies attributed the extensive degree of variability in this region to unequal meiotic exchanges involving the various classes of repetitive sequences in the centromeric region5,7,25 and to unequal exchanges involving stretches of homologous DNA sequences in the proximal short and long arms of chromosome 9.15 The results of the present study would support such an assumption. The microdissection-derived probes described here and elsewhere16 do not permit the discrimination between the 9p12 and 9q13-21.1 regions, and the origin and exact composition of the region 9p12/9q13-21.1 remains unclear. However, the enigma of the region 9p12/9q13-21.1 has to some extent been clarified by the observation that these regions are homologous to the short arms of the acrocentric chromosomes. Significantly, this study has also indicated the presence of breakpoint clustering in the regions stained by the microdissection-derived probe midi36.
References
Verma RS . A reply: pericentric inversion of chromosome 9qh are ‘real’ but the mechanisms of their origin are highly complex Hum Genet 1999 105: 183–184
Verma RS . Heterochromatin molecular and structural aspects New York: Cambridge University Press 1988 pp 276–299
Mitelman F . ISCN 1995: An international system for human cytogenetic nomenclature Basel: S. Karger 1995 pp 1–115
Gardner RJM, Sutherland GR . Chromosome abnormalities and genetic counseling In: Oxford monographs on medical genetics No. 29 New York and Oxford: Oxford University Press 1996 pp 139–158
Macera MJ, Verma RS, Conte RA, Bialer MG, Klein VR . Mechanisms of the origin of a G-positive band within the secondary constriction region of human chromosome 9 Cytogenet Cell Genet 1995 69: 235–239
Ramesh KH, Verma RS . Breakpoints in alpha, beta, and satellite III DNA sequences of chromosome 9 result in a variety of pericentric inversions J Med Genet 1996 33: 395–398
Samonte RV, Conte RA, Ramesh KH, Verma RS . Molecular cytogenetic characterization of breakpoints involving pericentric inversions of human chromosome 9 Hum Genet 1996 98: 576–580
Horvath JE, Bailey JA, Locke DP, Eichler EE . Lessons from the human genome: transitions between euchromatin and heterochromatin Hum Mol Genet 2001 10: 2215–2223
Senger G, Weimer J, Claussen U, Chuboda I . Microdissection and reverse chromosome painting In: Wegner RD (ed) Diagnostic Cytogenetics; Springer Lab Manual Heidelberg: Springer Verlag 1999 pp 356–375
Senger G, Chudoba I, Plesch A . Multicolor-FISH–the identification of chromosome aberrations by 24 colors BIOforum 1998 9: 499–503
Mrasek K, Heller A, Rubtsov N et al. Reconstruction of the female Gorilla gorilla karyotype by Zoo-FISH using 25-color FISH and multicolor banding (MCB) Cytogenet Cell Genet 93: 242–248
Liehr T, Thoma T, Kammler K et al. Direct preparation of uncultured EDTA-treated or heparinized blood for interphase FISH analysis Appl Cytogenet 1995 21: 185–188
Liehr T, Heller A, Starke H, Rubtsov N, Trifonov V, Mrasek K, Weise A, Kuechler A, Claussen U . Microdissection based high resolution multicolor banding for all 24 human chromosomes Int J Mol Med 2002 9: 335–339
Speicher MR, Gwyn Ballard S, Ward DC . Karyotyping human chromosomes by combinatorial multi-fluor FISH Nat Genet 1996 12: 368–375
Park JP, Wojiski SA, Spellman RA, Rhodes CH, Mohandes TK . Human chromosome 9 pericentric homologies: implications for chromosome 9 heteromorphisms Cytogenet Cell Genet 1998 82: 192–194
Müller-Navia J, Nebel A, Schleiermacher E . Complete and precise characterization of marker chromosomes by application of microdissection in prenatal diagnosis Hum Genet 1995 96: 661–667
Luke S, Verma RS . Genetic consequences of ‘euchromatic’ band within 9qh region Am J Med Genet 1993 45: 107
Hoo JJ, Szego K, Wong P, Roland B . Evidence of chromosome 9 origin of the euchromatic variant band within 9qh Clin Genet 1993 43: 309–311
Levy B, Dunn TM, Hirschhorn K, Kardon N . Jumping translocations in spontaneous abortions Cytogenet Cell Genet 2000 88: 25–29
Gebhart E, Liehr T . Patterns of genomic imbalances in human solid tumors (Review) Int J Onc 2000 16: 383–399
Bleichert A, Fiedler W, Claussen U et al. A long distance-PCR derived FISH probe detects a deletion between p15 and p16 in CML and T-ALL patients Int J Mol Med 2001 7: 591–595
Schinzel A . Catalogue of unbalanced chromosome aberrations in man Berlin, New York W. de Gruyter 2001 pp 1–966
Fryns JP, Kleczkowska A, Londers L, van den Berghe H . Unusual chromosome 9 polymorphism and reproductive failure Ann Genet 1985 28: 49–51
Wan TS, Ma SK, Chan LC . Acquired pericentric inversion of chromosome 9 in essential thrombocythemia Hum Genet 2000 106: 669–670
Wang JC, Miller WA . Molecular cytogenetic characterization of two types of chromosome 9 variants Cytogenet Cell Genet 1994 67: 190–192
Webb GC, Krumins EJ, Eichenbaum SZ, Voullaire LE, Earle E, Choo KH . Non C-banding variants in some normal families might be homogeneously staining regions Hum Genet 1989 82: 59–62
Reddy KS . Variants of chromosome 9 with additional euchromatic bands: two case reports Am J Med Genet 1996 64: 536–538
Knight LA, Soon GM, Tan M . Extra G positive band on the long arm of chromosome 9 J Med Genet 1993 30: 613
Jalal SM, Kukolich MK, Garcia M, Day DW . Euchromatic 9q+heteromorphism in a family Am J Med Genet 1990 37: 155–156
Hansmann I . Structural variability of human chromosome 9 in relation to its evolution Hum Genet 1976 31: 247–262
Madan K . An extra band in human 9qh+ chromosomes Hum Genet 1978 43: 259–264
Berg JM, Gardner HA, Gardner RJ et al. Dic(21;21) in a Down's syndrome child with an unusual chromosome 9 variant in the mother J Med Genet 1980 17: 144–148
Donlon TA, Magenis RE . Structural organization of the heterochromatic region of human chromosome 9 Chromosoma 1981 84: 353–363
Silengo MC, Davi GF, Franceschini P . Extra band in the 9qh+ chromosome in a normal father and in his child with multiple congenital anomalies Hum Genet 1982 60: 294
Docherty Z, Hulten MA . Extra euchromatic band in the qh region of chromosome 9 J Med Genet 1985 22: 156–157
Wojiski SA, Rhodes CH, Brodhurst CA, Mohandas TK, Park JP . The G positive band of the rare euchromatic 9qh variant is derived from 9p12 Appl Cytogenet 1997 23: 125–129
Luke S, Verma RS, PeBenito R, Macera MJ . Inversion-duplication of bands q13–q21 of human chromosome 9 Am J Med Genet 1991 40: 57–60
Madan K, Bobrow M . Structural variation in chromosome no. 9 Ann Génét 1974 17: 81–86
Verma RS, Luke S, Brennan JP, Mathews T, Conte RA, Macera MJ . Molecular topography of the secondary constriction region (qh) of human chromosome 9 with an unusual euchromatic band Am J Hum Genet 1993 52: 981–986
Colls P, Blanco J, Martinez-Pasarell O et al. Chromosome segregation in a man heterozygous for a pericentric inversion, inv(9)(p11q13), analyzed by using sperm karyotyping and two-color fluorescence in situ hybridization on sperm nuclei Hum Genet 1997 99: 761–765
Sasagawa I, Ishigooka M, Kubota Y, Tomaru M, Hashimoto T, Nakada T . Pericentric inversion of chromosome 9 in infertile men Int Urol Nephrol 1998 30: 203–207
Cora T, Demirel S, Acar A . Chromosomal abnormalities in mentally retarded children in the Konya region–Turkey Genet Couns 2000 11: 53–55
Miyaoka T, Seno H, Itoga M, Ishino H . A case of small cerebral cyst and pericentric inversion of chromosome 9 that developed schizophrenia-like psychosis Psychiatry Clin Neurosci 1999 53: 599–602
Zenker M, Dorr HG . A case associated with Walker Warburg syndrome phenotype and homozygous pericentric inversion 9: coincidental finding or aetiological factor? Acta Paediatr 2000 89: 750–751
Stanojevic M, Stipoljev F, Koprcina B, Kurjak A . Oculo-auriculo-vertebral (Goldenhar) spectrum associated with pericentric inversion 9: coincidental findings or etiologic factor? J Craniofac Genet Dev Biol 2000 20: 150–154
Milan C, Lamberti L . Study of C-polymorphisms of chromosomes 1, 9 and 16 in lymphocytes of patients with laryngeal carcinoma Boll Soc Ital Biol Sper 1996 72: 187–194
Jackson MS, Rocchi M, Thompson G et al. Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications and unstable sequences with homologies to telomeric and other centromeric locations Hum Mol Genet 1999 8: 205–215
Acknowledgements
Supported in part by the Herbert Quandt Stiftung der VARTA AG, the Madeleine Schickedanz-Kinderkrebs-Stiftung, the Wilhelm Sander-Stiftung (99.105.1) and the EU (ICA2-CT-2000-10012 and QLRT-1999-31590). The continued support from the Carl Zeiss GmbH, Jena, is gratefully acknowledged. The authors thank Dr H-Ulli Weier (Berkeley, USA) and Dr Lyndal Kearney (London, UK) for very helpful suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Starke, H., Seidel, J., Henn, W. et al. Homologous sequences at human chromosome 9 bands p12 and q13-21.1 are involved in different patterns of pericentric rearrangements. Eur J Hum Genet 10, 790–800 (2002). https://doi.org/10.1038/sj.ejhg.5200889
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200889
Keywords
This article is cited by
-
Acrocentric Chromosome Polymorphic Variants on Chinese Female Have Possible Association with Unexplained Recurrent Pregnancy Loss
Reproductive Sciences (2021)
-
Pericentric inversion (Inv) 9 variant—reproductive risk factor or benign finding?
Journal of Assisted Reproduction and Genetics (2019)
-
Analyses of karyotype by G-banding and high-resolution microarrays in a gender dysphoria population
Genes & Genomics (2018)
-
Molecular characterization and evaluation of complex rearrangements in a case of ring chromosome 15
Molecular Cytogenetics (2017)
-
Insulin-like growth factor type 1 deficiency in a Moroccan patient with de novo inverted duplication 9p24p12 and developmental delay: a case report
Journal of Medical Case Reports (2016)