Abstract
Polypeptide chain initiation factor eIF4GI undergoes caspase-mediated degradation during apoptosis to give characteristic fragments. The most prominent of these has an estimated mass of approximately 76 kDa (Middle-Fragment of Apoptotic cleavage of eIF4G; M-FAG). Subcellular fractionation of the BJAB lymphoma cell line after induction of apoptosis indicates that M-FAG occurs in both ribosome-bound and soluble forms. Affinity chromatography on m7GTP-Sepharose shows that M-FAG retains the ability of eIF4GI to associate with both the mRNA cap-binding protein eIF4E and initiation factor eIF4A and that the ribosome-bound form of M-FAG is also present as a complex with eIF4E and eIF4A. These data suggest that the binding sites for eIF4E, eIF4A and eIF3 on eIF4GI are retained in the caspase-generated fragment. M-FAG is also a substrate for cleavage by the Foot-and-Mouth-Disease Virus-encoded L protease. These properties, together with the pattern of recognition by a panel of antibodies, define the origin of the apoptotic cleavage fragment. N-terminal sequencing of the products of caspase-3-mediated eIF4GI cleavage has identified the major cleavage sites. The pattern of eIF4GI degradation and the possible roles of the individual cleavage products in cells undergoing apoptosis are discussed. Cell Death and Differentiation (2000) 7, 628–636
Similar content being viewed by others
Introduction
In eukaryotic cells the polypeptide chain initiation factor eIF4G plays an essential role in the mechanism of translation by acting as a molecular bridge between other components of the ribosomal initiation complex (reviewed in references1,2,3). Two forms of eIF4G, which are products of different genes, have been identified.4 eIF4GI is a single polypeptide chain of at least 154 kDa;5 recently extensions to the original N-terminal sequence of this protein have been described,6,7,8 and it is possible that the true N-terminus has not yet been identified. In vivo eIF4G exists partly in the form of a complex with the mRNA cap-binding protein eIF4E and the ATP-dependent RNA helicase eIF4A, constituting the initiation factor eIF4F.9 Within the sequence of eIF4GI9 there are domains that interact with eIF4E,10 eIF4A,10,11, eIF3,11 the poly(A) binding protein (PABP)7,12,13,14 and the eIF4E kinase Mnk1.15 Interaction of PABP with eIF4GI has been suggested to facilitate the functional association of the 3′ end of an mRNA with the 5′ end.14 The association of eIF4G with eIF4E markedly enhances the binding of the latter to the mRNA cap16 and is necessary to permit cap-dependent translation. It is also likely that the phosphorylation of eIF4E, which has been correlated with enhanced translational activity in cells treated with mitogens and growth factors,17,18,19,20 is facilitated by the binding of the eIF4E kinase Mnk1 to the C-terminal part of eIF4GI.15 Over-expression of either eIF4G or eIF4E has been shown to transform cells to a tumorigenic phenotype,21,22 and abnormally high levels of both factors have been observed in several types of human tumours.23,24,25
There is a well-characterised loss of integrity of both eIF4GI (reviewed in reference2) and eIF4GII4,26 in mammalian cells after infection with picornaviruses such as Polio or Foot-and-Mouth-Disease viruses (FMDV). This is a result of cleavage at specific internal sites by the virally-encoded 2A and L proteases, respectively. Since these cleavages occur in a region between the eIF4E and eIF3 binding sites27 they destroy the bridging function of eIF4G and thus lead to the inhibition of cap-dependent initiation.28,29,30,31,32 The remaining C-terminal fragment of eIF4G can nevertheless still support initiation that is cap-independent28 or that involves ribosome binding to internal sites on mRNAs.33
We have shown that eIF4GI is rapidly cleaved during the process of apoptosis (cell death) that can be induced by a variety of treatments in several cell lines.34,35,36 This effect is selective since several other initiation factors, including eIF4E and eIF4A, are relatively stable under these conditions. In cells undergoing apoptosis proteolysis of eIF4GI to produce characteristic cleavage fragments is mediated by one or more caspases,34 and caspase-3 activity is both necessary and sufficient for this process.36,37 Protein synthesis is strongly inhibited in cells undergoing apoptosis and in some situations this inhibition, as well as the cleavage of eIF4GI, can be prevented by incubation with the cell-permeable caspase inhibitor, z-VAD.FMK.34,35,36 It is possible that the down-regulation of translation in apoptotic cells is due to the specific and complete cleavage of eIF4G. However the remaining protein synthetic activity (which amounts to some 35% of the control rate35) may be maintained by the continued presence of one or more cleavage fragments that retain the ability to interact with other key initiation factors. If this is the case it is also likely that the properties of the cleavage fragments that appear during apoptosis are distinct from those of the fragments of eIF4G produced in cells during picornavirus infection, which do not support any cap-dependent initiation.28,29,30,31,32
To begin to test these hypotheses we have identified the sites of the caspase-mediated cleavages of eIF4GI that occur rapidly during cellular apoptosis and have characterised the properties of the major cleavage products (N-FAG, M-FAG and C-FAG) that persist in cells for several hours after fragmentation of the full-length factor. Our results indicate that the cleavages that occur and the nature of the fragments that result, are quite distinct from those seen in picornavirus-infected cells. The properties of the M-FAG fragment observed in apoptotic cells are compatible with the possibility that this protein can still support cap-dependent initiation but with a substantially impaired efficiency.
Results
Characterisation of the cleavage fragments of eIF4G in apoptotic cells
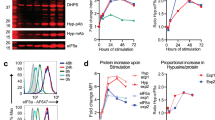
Treatment of the EBV-negative Burkitt's lymphoma BJAB cell line with agents such as anti-Fas (CD95) antibody, etoposide and cycloheximide induces apoptosis. In this system cycloheximide is the most potent and rapidly acting of the inducers examined, as revealed by FACS analysis of the extent of accumulation of cells with a less than G0/G1 DNA content (Figure 1A). We have reported previously that all of these treatments also result in the complete cleavage of eIF4GI into a series of characteristic fragments.34 Figure 1B illustrates the time-course of eIF4GI cleavage in response to induction of apoptosis with cycloheximide, as visualised with a number of domain and sequence-specific antisera (W, RL, E; see Materials and Methods and Figure 3). In agreement with our previous data,34,35 antiserum generated against a large C-terminal fragment of eIF4G (W) showed that cycloheximide induced the cleavage of total cytoplasmic eIF4GI to yield the novel ca. 76 kDa fragment (M-FAG; Figure 1B, lanes 4–6). Cleavage of eIF4GI correlated temporally with that of the classical apoptotic substrate, poly(ADP-ribose) polymerase (PARP; lanes 4–6). M-FAG is actually a doublet, the levels of both components of which remain constant within cells for several hours after induction of apoptosis. In addition, with this antiserum we have observed the transient generation of smaller amounts of eIF4GI fragments of approximately 150 kDa (p150, see Figure 2A) and 120 kDa (p120, faintly visible in Figure 1B and 2A), which appear within 2 h of cycloheximide treatment. The N-terminal sequence-specific antiserum (RL) visualised a group of previously described cleavage products (eIF4GcpN),36 which we designate N-FAG, and the C-terminal sequence-specific antiserum (E) recognised a novel fragment of approximately 50 kDa (C-FAG) (Figure 1B). The same pattern of cleavages was observed after treatment with other inducers of apoptosis (serum deprivation, anti-Fas antiserum and etoposide – data not shown). Similar results were also seen using antisera specific to eIF4GII, although in this case a different pattern of cleavages occurred.38
Induction of apoptosis in BJAB cells: Time-course of eIF4GI cleavage and analysis of the ability of the cleavage fragments to interact with eIF4E. (A) Determination of the extent of apoptosis by FACS analysis of DNA content. Cells were incubated without or with anti-Fas antibody (50 ng/ml) or etoposide (100 μg/ml) for 24 h or with cycloheximide (100 μg/ml) for 12 h. They were then stained with propidium iodide and their DNA content determined by FACS analysis as described previously.34 Regions of each profile corresponding to cells in the G0/G1 or S/G2 phases of the cell cycle and to apoptotic cells with a sub-G1 DNA content (labelled ‘A’) are indicated. (B) Exponentially growing cells were treated with cycloheximide for the times shown and cytoplasmic extracts prepared as described in the Materials and Methods. Equal quantities of protein from each extract were subjected to SDS gel electrophoresis, followed by immunoblotting for eIF4GI (with antiserum W, RL, or E), eIF4A, eIF4E, the phosphorylated form of eIF4E and PARP, as indicated. Molecular mass markers and the locations of the group of N-terminal fragments (N-FAG), the middle fragment (M-FAG), and the C-terminal (C-FAG) fragment of eIF4GI (see Figure 3A), together with a faintly stained transient intermediate (p120), are indicated. These data are from a single experiment but are representative of those obtained on five separate occasions. Note that the low molecular mass bands in lanes 1–4 which are recognised by antiserum RL are cross-reacting proteins which are distinct from the N-FAG fragments. (C) Aliquots of cell extracts containing equal amounts of total cytoplasmic protein, prepared as above, were subjected to affinity chromatography on m7GTP-Sepharose to purify eIF4E and associated proteins. Recovered protein was resolved by SDS gel electrophoresis and immunoblotted for eIF4GI and its cleavage fragments (using antisera RL and E), eIF4A, eIF4E and the phosphorylated form of eIF4E, as indicated. These data are from a single experiment but are representative of those obtained on five separate occasions
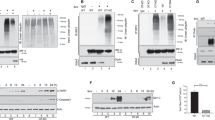
Localization of the caspase-3 cleavage sites of eIF4GI by immunoblotting. (A) A diagrammatic representation of the major structural domains of eIF4GI is shown, indicating the sites of interaction with PABP, eIF4E, eIF3 and eIF4A, the L protease cleavage site and regions of sequence used to generate specific antisera (see Materials and Methods). In addition, the two major caspase-3 cleavage sites, which lead to the generation of N-FAG, M-FAG and C-FAG are indicated (see text for details). (B) BJAB cells were incubated in the absence (lane 1) or presence (lane 2) of anti-Fas antiserum (500 ng/ml) for 4 h. Extracts were prepared and resolved by electrophoresis on 7.5% SDS gels. The gels were then immunoblotted with antiserum specific for eIF4GI (antiserum W). Purified FLAG-eIF4GI was incubated in the absence (lanes 3, 5, 7, 9, 11 and 13) or presence (lanes 4, 6, 8, 10, 12 and 14) of recombinant caspase-3 as described in the Materials and Methods. eIF4GI and its cleavage fragments were resolved by gel electrophoresis on 7.5% gels and identified by immunoblotting using the following domain-specific and sequence-specific antisera: lanes 3–6; W; lanes 7 and 8; H; lanes 9 and 10; E; lanes 11 and 12; RL; lanes 13 and 14; B. (C) Purified FLAG-eIF4GI was incubated in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of recombinant caspase-3 as described in (B). eIF4GI and its cleavage fragments were then resolved by electrophoresis on 12.5% SDS gels and visualised by immunoblotting using antiserum E (lanes 1 and 2) or W (lanes 3 and 4). Molecular mass markers and the positions of the major cleavage fragments of eIF4GI are indicated. Note that the low molecular mass band stained with antiserum E in lane 1 of (C) is a cross-reacting species which is distinct from C-FAG. These data are from a single experiment but are representative of those obtained on four separate occasions
The p150, p120 and M-FAG cleavage fragments of eIF4GI are associated with ribosomes and are distinct from those generated by L protease. (A) Cells were incubated in the absence (lanes 1, 3, 5) or presence (lanes 2, 4, 6) of 100 μg/ml cycloheximide for 4 h and cytoplasmic extracts prepared. Equal quantities of protein were either analysed directly (lanes 1 and 2), or used to prepare post-ribosomal supernatant (lanes 3 and 4) or ribosomes (lanes 5 and 6), as described in the Materials and Methods. Proteins were analyzed by SDS gel electrophoresis and immunoblotting with antiserum specific for eIF4GI (antiserum W). (B) Equal amounts of cell extracts (lanes 1 and 2), post-ribosomal supernatants (lanes 3 and 4) or ribosomes (lanes 5 and 6) were subjected to affinity chromatography on m7GTP-Sepharose to purify eIF4E and associated proteins. Recovered protein was resolved by SDS gel electrophoresis and immunoblotted for eIF4GI using antiserum W. (C) Reticulocyte lysate was incubated in the absence (lane 1) or presence of caspase-3 (10 μg/ml) (lane 2) or L protease (0.3 μg/ml) (lanes 3 and 4) for 30 min at 30°C, prior to the addition of buffer (lanes 1–3) or caspase-3 (lane 4) for a further 30 min. Aliquots of lysate were analyzed by SDS gel electrophoresis and immunoblotting with antiserum specific for eIF4GI (antiserum W). In each panel molecular mass markers and the positions of the major cleavage fragments of eIF4GI are indicated. The diamond in (C) identifies a novel L protease- and caspase-3-generated fragment of eIF4GI. These data are from a single experiment but are representative of those obtained on three separate occasions
We also assessed the levels of several other initiation factors in the BJAB cells under the same conditions. Figure 1B shows that, in agreement with our previous results,34 the levels and integrity of poly(A)-binding protein (PABP), eIF4A and total eIF4E were unaffected in apoptotic cells. However, use of antiserum which was specific for the phosphorylated form of eIF4E39 indicated that the phosphorylation status of eIF4E decreased at later times following the induction of apoptosis (Figure 1B, lanes 4–6).
Association of the cleavage fragments of eIF4G with eIF4E and ribosomes
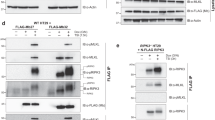
Initiation factor eIF4E and associated proteins were isolated by m7GTP-Sepharose chromatography (Figure 1C). Immunoblotting of the m7GTP-Sepharose-purified proteins revealed that M-FAG (lanes 4–6), p120 (lanes 4, 5) and eIF4A (lanes 4–6) remained associated with eIF4E in cells in which essentially all of the eIF4GI had been cleaved. However, when protein recovered by m7GTP-Sepharose was visualised with antisera specific for N-FAG or C-FAG, these characteristic fragments were absent, indicating that neither of these products remains associated with eIF4E (Figure 1C, lanes 4–6). Consistent with this and the location of the site of interaction of PABP with eIF4GI in the N-terminal domain of the latter protein,6,7 the level of PABP recovered using m7GTP-Sepharose chromatography was decreased following the induction of apoptosis (data not shown). These data suggest that M-FAG retains binding sites for eIF4E and eIF4A but has lost the N-terminal and C-terminal regions of eIF4GI.
A significant proportion of eIF4GI is bound to ribosomes in mammalian cells.40 This is believed to be a consequence of the association of eIF4GI with eIF3 which is attached to native ribosomal subunits.11 We have investigated whether the cleavage fragments of eIF4GI retain the ability to bind to ribosomes in apoptotic cells. Figure 2A shows that although some M-FAG was found in the post-ribosomal supernatant of apoptotic cells (lane 4), a substantial proportion of this fragment was retained by the ribosomes (lane 6). Similarly, both the p150 and p120 fragments of eIF4GI were recovered with the ribosomal fraction (lane 6) whether apoptosis was induced with cycloheximide (Figure 2A) or etoposide (data not shown). Thus it is likely that these fragments also retain the domain that is required for the association of eIF4GI with eIF3 (and hence the ribosome) in vivo. To address whether these ribosomal-bound fragments of eIF4GI further possess the ability to interact with eIF4E, we have isolated eIF4E and associated proteins from these fractions by m7GTP-Sepharose chromatography. Analysis by immunoblotting of total (Figure 2B, lanes 1, 2), ribosome-free (lanes 3, 4) and ribosome-bound material (lanes 5, 6) showed the presence of some p150 (faint band in lane 6), p120 (lanes 2, 4, 6) and M-FAG fragments of eIF4G (lanes 2, 4, 6) associated with eIF4E in extracts from apoptotic cells.
Identification of the caspase-3 cleavage sites in eIF4G and comparison with picornavirus protease-mediated cleavage
We have compared the caspase-3-generated eIF4GI cleavage products with the fragments produced by the site-specific L protease from FMDV (Figure 2C). Incubation of a reticulocyte lysate with L protease generated the characteristic cleavage fragment, Ct, which lacks the eIF4E binding site28,41 and is distinct from M-FAG (compare lane 3 with lane 1). Exposure to caspase-3 alone generated p120 and M-FAG (lane 2 versus lane 1) while incubation of reticulocyte lysate with L protease followed by caspase-3 led to the further cleavage of Ct to give an even smaller product (lane 4 versus lane 3). Thus Ct remains sensitive to caspase attack. Incubation of extracts from apoptotic cells with L protease in vitro showed that the viral enzyme degraded not only full-length eIF4GI to produce the Ct fragment, but further processed the p120 and M-FAG fragments, resulting in loss of binding to eIF4E (data not shown). In addition, immunoblots using antiserum RL demonstrated that the N-terminal fragment of eIF4G released by poliovirus 2A protease cleavage between residues 642 and 643 (using the numbering system of Imataka et al.7) could also serve as a substrate for caspase-3 (data not shown). Together these data indicate that p120 and M-FAG can bind to both eIF4E and ribosomes and that the specific sites of L and 2A protease cleavage are distinct from those utilized by caspase-3.
Previously we have shown that caspase-3 is both necessary and sufficient for the cleavage of eIF4GI.37 To determine the sites of cleavage of eIF4GI by this enzyme we have incubated purified, baculovirus-expressed FLAG-eIF4GI with caspase-3 in vitro and analyzed the resulting fragments by immunoblotting with sequence-specific antisera followed by mass spectrometry and protein sequencing. Figure 3A shows a domain map of eIF4GI with the binding sites for eIF4E, eIF3, eIF4A and PABP, indicating the regions to which antisera were raised. These antisera were directed against large regions of the C-terminus (W) or the N-terminus (RL) of eIF4GI, as described in Materials and Methods, and against three peptide epitopes (B, H and E). In agreement with our previous findings,37 cleavage of eIF4GI by caspase-3 in vitro generated fragments with the same mobility on SDS-gel electrophoresis as those observed in apoptotic (anti-Fas-stimulated) cells (Figure 3B, compare lanes 2 and 4). The immunoblots in Figure 3B (lanes 5–14) show that all the antisera recognised intact eIF4GI in control and caspase-3-treated preparations of FLAG-eIF4GI. The M-FAG doublet that appeared after incubation with caspase-3 was decorated only by antisera W (lane 6) and H (lane 8). Neither component of the doublet reacted with antiserum E (lane 10), antiserum RL (lane 12) or antiserum B (lane 14). The p150 fragment was recognised by antisera W (Figures 2 and 3B, lane 6), H (Figure 3B, lane 8), RL (Figure 3B, lane 12) and weakly by antiserum B (Figure 3B, lane 14). In addition, the antisera RL (Figure 3B, lane 12) and B (Figure 3B, lane 14) recognised a group of bands (N-FAG; see Figure 1) which correspond to the N-terminal fragment of eIF4GI (eIF4GcpN).36 These results, and those presented in Figures 1 and 2, are consistent with the conclusion that the M-FAG fragment arises from cleavage of eIF4GI at two sites and retains the eIF4E and eIF3 binding sites (Figure 3A). The p120 and p150 fragments represent products from a single caspase-3 cleavage event in the N-terminal and C-terminal halves of eIF4GI, respectively, and are thus alternative intermediates in the generation of M-FAG (see Figure 1).
To determine precisely the cleavage sites in eIF4GI we have subjected the M-FAG cleavage fragment to N-terminal microsequencing. Purified FLAG-eIF4GI was cleaved in vitro with caspase-3, subjected to SDS gel electrophoresis and blotted on to PVDF membrane prior to protein sequencing of the M-FAG fragment. The sequence obtained, AFKEANPA(D)VPE (where the brackets indicate ambiguity in the residues identified), corresponds to a region in the deduced amino acid sequence of human eIF4GI, comprising amino acids 493–503 (based on the numbering system employed by Imataka et al.;7 accession number AF104913). This indicates that caspase-3 cleavage had occurred on the C-terminal side of the sequence DLLD, between residues 492 and 493. To identify the cleavage site at the C-terminus of eIF4GI, fragments were resolved on a higher percentage acrylamide gel and C-FAG was identified with antisera E (Figure 3C, lane 2) and W (Figure 3C, lane 4). Additionally, C-FAG derived from caspase-3 cleavage of native eIF4GI in eIF4F preparations was gel purified. Subsequent protein microsequencing, together with analysis by mass spectrometry (data not shown), gave the sequence R(M)AR(G)TPATKRS. This establishes the second major caspase-3 cleavage site as being on the C-terminal side of the sequence DRLD, between residues 1136 and 1137. The calculated Mr of the sequence that lies between amino acids 493 and 1136 of eIFGI is 72244. This agrees well with the estimated Mr of 76 kDa for M-FAG, which was obtained by comparison with the migration of molecular mass markers on SDS gel electrophoresis.
Discussion
The work described here sheds further light on the nature of the changes undergone by initiation factor eIF4GI during apoptosis.42 We have previously shown that cells maintain a residual rate of protein synthesis for up to 2 days following induction of apoptosis in response to serum deprivation, treatment with anti-Fas antibody or exposure to etoposide.34,35 The data indicate that, although full-length eIF4GI is rapidly lost from apoptosing cells, three relatively long-lasting cleavage fragments are generated, one of which (M-FAG) retains the ability to interact with other key initiation factors. Our results suggest that M-FAG may continue to permit some cap-dependent translation (albeit perhaps inefficiently) since it still interacts with eIF4E, eIF4A and ribosomes. Both the structure and the functional properties of the cleavage fragments of eIF4GI that appear during apoptosis are distinct from those of the fragments produced in cells during picornavirus infection.28,29,30,31,32
Using a panel of eIF4GI-specific antibodies (Figure 3), together with direct N-terminal protein sequencing and mass spectrometry analysis (data not shown), we have established that the M-FAG fragment is generated from caspase-mediated cleavages between amino acids 492 and 493 at the N-terminus and amino acids 1136 and 1137 at the C-terminus. The sites of proteolysis lie immediately downstream of the sequences DLLD and DRLD respectively and are consistent with the known site preferences of caspase-3 for DXXD motifs.43 It is possible that some cleavage also occurs after the sequence DRGD (residues 1120/1121 or 1133/1134), giving rise to the doublet pattern of M-FAG observed on blots (Figures 1,2,3). However there are other DXXD sequences in eIFGI which do not appear to be targeted by caspase-3, perhaps because they are inaccessible to the enzyme.
The ability of M-FAG to bind to eIF4E and ribosomes is consistent with the identified cleavage sites since the domains that interact with eIF4E, eIF4A and eIF3 remain in this fragment (Figure 3). Although one eIF4A binding site in the C-terminal part of eIF4G is absent from M-FAG (Figure 3), eIF4A is still present in the m7GTP-Sepharose-purified fraction from apoptotic cells (Figure 1C), probably because it can also bind to the central region of eIF4GI.44 In contrast, M-FAG is predicted not to bind to PABP since the latter interacts with the N-terminus of eIF4GI.6,7 We have shown previously that the PABP which is present in an m7GTP-Sepharose-purified fraction from control Jurkat cells is in fact selectively lost from this fraction after induction of apoptosis.34 Nevertheless, in the present study, a small amount of PABP remained associated with eIF4E in apoptotic BJAB cells at a time when eIF4G was cleaved completely, suggesting that PABP may also interact with another eIF4E-associated factor.
Taken together these data suggest that a new form of eIF4F, containing eIF4E and eIF4A but with M-FAG in place of full-length eIF4GI, is associated with the ribosomes in apoptotic cells (Figure 4). This novel complex appears at about the time that the rate of translation is decreased.34,35 Unlike the situation in picornavirus-infected cells, where viral protease-mediated cleavage of eIF4G separates the domain that interacts with eIF4E from other functional regions of the protein,11,27,28,41 the modified form of eIF4F that arises in apoptotic cells may still be able to support at least some cap-dependent initiation. Imataka and Sonenberg44 have shown that both binding sites for eIF4A may be required for stimulating translation and it is possible therefore that the M-FAG-containing eIF4F may be less efficient in mRNA unwinding activity. Nevertheless, two reports45,46 have recently shown that a central core domain of eIF4G can still act in the recruitment of ribosomes to mRNA, in the absence of the C-terminal eIF4A binding site.
Model for the function of eIF4GI in initiation before and after caspase-mediated cleavage in apoptotic cells. (A) Domain structures of intact eIF4GI and the cleavage fragment M-FAG. Shaded regions indicate the sites of interaction with PABP, eIF3, eIF4A and the eIF4E kinase Mnk1. The filled region indicates the site of interaction with eIF4E. Note that binding sites for eIF4E, eIF3 and eIF4A are retained in M-FAG but that the sites of interaction with PABP and Mnk1 are lost. (B) Components of the (40S ribosome.mRNA) initiation complex in non-apoptotic cells. The complex interacts with both the 5′ cap and 3′ poly(A) tail of the mRNA molecule through eIF4E (labelled E) and PABP (labelled P) respectively. (C) Proposed structure of the (40S ribosome.mRNA) initiation complex in apoptotic cells. The complex, which contains M-FAG in place of intact eIF4GI, continues to interact with the mRNA cap structure via eIF4E but is impaired in its ability to bind PABP and hence the poly(A) tail. The poly(A) tract may remain associated with PABP and the N-FAG fragment of eIF4GI
The M-FAG-containing form of eIF4F is also likely to be deficient in PABP-dependent re-initiation activity since the ability to cause circularisation of mRNA as a result of simultaneous interaction of PABP with poly(A) (at the 3′ end) and of eIF4E with the cap structure (at the 5′ end)7,12,14 will be reduced (see Figure 4). Furthermore, we have observed that the phosphorylation state of eIF4E also decreases in BJAB cells following the induction of apoptosis (Figure 1). This may be an additional consequence of eIF4G cleavage in that the ability of the eIF4E kinase, Mnk1, to phosphorylate its substrate is likely to be compromised as a result of the loss of the proximity of Mnk1 to eIF4E mediated by eIF4G bridging.15 As discussed elsewhere,42 these several changes in the properties of eIF4G could constitute the basis for the decrease in the rate of overall protein synthesis observed in apoptotic cells.34,35,36
Another possibility is that one or more of the eIF4G cleavage fragments that accumulate in apoptotic cells may have a dominant-negative effect with respect to the functions of the intact factor. Since PABP binds to the N-terminus of eIF4GI7 the N-FAG fragment may impair the process of poly(A)-dependent initiation, or its regulation, by sequestering PABP (Figure 4). C-FAG may similarly interfere with eIF4E phosphorylation by binding Mnk1. A fragment of a gene that encodes a mammalian homologue of the C-terminal part of eIF4GI, variously called p97, DAP-5 or NAT1,47,48,49 has been reported to protect HeLa cells against interferon-γ-induced apoptosis. Whereas p97/DAP-5/NAT1 is distinct from M-FAG in that the former lacks the eIF4E binding site, both M-FAG and C-FAG have some sequence similarities to p97/DAP-5/NAT1 and one or both may exert similar functions.42 Further work is underway to determine the functions of N-FAG, M-FAG and C-FAG in the regulation of translation, to characterise the M-FAG-containing eIF4F complex and to address the possible role of this complex in mediating cap-dependent and/or -independent translation in apoptotic cells.
Materials and Methods
Materials
Tissue culture materials were purchased from Life Technologies; cycloheximide was obtained from Sigma-Aldrich; purified caspase-3 was purchased from Autogen Bioclear (UK); agonistic anti-Fas antibody (clone CH-11) was provided by Upstate Biotechnology; antiserum specific to poly(ADP-ribose) polymerase (PARP) was from Boehringer Mannheim.
Cell culture and induction of apoptosis
The EBV-negative Burkitt's lymphoma cell line BJAB was cultured as described by Clemens et al.34 Apoptosis was induced in BJAB cells by exposure to cycloheximide (100 μg/ml), anti-Fas antibody (500 ng/ml) or etoposide (100 μg/ml) for the times indicated in the individual Figure legends. The extent of apoptosis was monitored by the proportion of cells with less than a G0/G1 content of DNA, as determined by FACS analysis, and by the cleavage of PARP.34
Preparation and analysis of cell extracts
Cytoplasmic extracts of cells were prepared as described previously.34 Initiation factor eIF4E and associated proteins were isolated by affinity chromatography on m7GTP-Sepharose.17,35,50,51 Samples were subjected to electrophoresis on SDS polyacrylamide gels and the proteins transferred to PVDF membranes (Millipore) using a semi-dry blotting apparatus (Hoefer). Equal amounts of total protein or fractions purified on m7GTP-Sepharose from equal amounts of cell extract were loaded on the gels. Ribosomal and post-ribosomal fractions of extracts from control and apoptotic cells were prepared as described previously.40
Antisera
Antibodies to eIF4E, eIF4A and the C-terminal region of eIF4GI33 (amino acids 1079–1560 using the numbering system of Imataka et al.7 (antiserum W)) were as described previously.34,35,41 Additional antisera specific for eIF4GI were developed in rabbits using the following peptides: KKEAVGDLLDAFKEVN (amino acids 483–498) (antiserum B); KKQKEMDEAATAEERERLKEELEEAR (amino acids 824–849) (antiserum H); RTPATKRTFSKEVEERSRERPSQPEGLR (amino acids 1139–1166) (antiserum E). Antiserum specific to the N-terminus of eIF4GI (antiserum RL) was described previously.36,52 Immunoblots were developed and quantified as in earlier studies.34,35
Preparation of recombinant proteins
1×106 plaques from a HeLa cell cDNA expression library (Stratagene) were screened by hybridization with a BglII DNA fragment (labelled by random priming) spanning the first 700 nucleotides of plasmid p4/16.6 Positive clones were recovered and were further analyzed by PCR with primers specific for the eIF4GI gene and the bacteriophage T3 promoter. PCR fragments were purified on Quiaquick (QIAGEN) columns and sequenced. Two clones (8/1 and 2/1) containing the longest 5′ end inserts were isolated by three successive rounds of plating and screening by PCR. Plasmids were then rescued from lambda phage infected cells by in vivo excision using the ExAssist/SOLR system (Stratagene). Analysis of the 3′ end sequences showed that they encoded eIF4GI (up to nucleotides 1135 for 2/1 and 1556 for 8/1). Although the 5′ sequences overlapped with the p4/16 insert,6 the 8/1 (accession number AF002815) and 2/1 (accession number AF002816) clones were distinct at their 5′ ends. The reading frames deduced from these sequences were in frame with the published eIF4GI amino acid sequence and with the p4/16 open reading frame, but they were each different at their N termini and lacked an identifiable 5′ non-coding sequence. The DNA sequence was confirmed twice for each clone by sequencing both strands.
To express eIF4GI in baculovirus the multiple cloning site of the baculovirus transfer vector pBackPac (Clontech) was modified by insertion of two annealed oligonucleotides: (gatccACCATGGACTACAAGGACGACGATGACAAGCCCGGGATACGAATTCGAC and tcgaGTCGAATTCGTATCCCGGGCTTGTCATCGTCGTCCTTGTAGTCCATGGTg) between the BamHI and XhoI sites. The resulting plasmid (pBac-pak His2) contained the FLAG epitope sequence followed by XmaI and EcoRI sites. The 5′ terminal XmaI–EcoRI fragment of p8/1, followed by the EcoRI insert of plasmid pHFC1,5 were successively subcloned in pBac-pak His2. The entire eIF4GI sequence fused to the FLAG epitope was purified after complete digestion with XhoI and partial digestion with BamHI and then subcloned into a second baculovirus shuttle vector, pFASTbac (Life Technologies). The pFASTbac-FLAG-eIF4GI was then introduced into the E. coli strain (DH10bac) containing the Bacmid and recombinant baculovirus was obtained following the manufacturer's instructions (Life Technologies). FLAG-eIF4GI was purified from Sf9 insect cells infected for 72 h with the recombinant baculovirus. Cells were harvested and resuspended in 5 ml per 100 ml culture of lysis buffer (50 mM MOPS, pH 7.2, 100 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1 mM phenylmethylsulphonyl fluoride and 1 mM benzamidine), lysed by addition of IGEPAL (Sigma) to 1% (by volume) and clarified by centrifugation at 15 000×g for 20 min at 4°C. The supernatant was applied to anti-FLAG-M2 affinity gel (Anachem, UK), washed in Buffer A (20 mM MOPS, pH 7.2, 50 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1 mM phenylmethylsulphonyl fluoride and 1 mM benzamidine) and stored at −80°C in aliquots. Recombinant Foot-and-Mouth-Disease Virus L protease was prepared as described previously.28,40,41
Determination of cleavage sites in eIF4GI
FLAG-eIF4GI (in Buffer A) was incubated with recombinant caspase-3 (10 μg/ml) for 30 min at 37°C, as described in the Figure legends. Samples were resolved by SDS–PAGE and proteins transferred to PVDF. Following identification of the cleavage fragments in adjacent tracks by immunoblotting, bands were excised and subjected to mass spectrometry and N-terminal sequencing using in-house facilities at the University of Sussex and the University of Oklahoma Health Sciences Center.
Abbreviations
- Ct:
-
C-terminal fragment of eIF4G
- eIFs:
-
eukaryotic initiation factors
- FAG:
-
fragment of the apoptotic cleavage of eIF4G
- FMDV:
-
foot-and-mouth-disease virus
- m7GTP:
-
7-methylguanosine triphosphate
- PABP:
-
poly(A) binding protein
- PARP:
-
poly(ADP-ribose) polymerase
- z-VAD.FMK:
-
Z-Val-Ala-Asp.fluo-romethylketone
References
Merrick WC and Hershey JWB . (1996) The pathway and mechanism of protein synthesis. In: Translational control, Hershey JWB, Mathews MB and Sonenberg N, eds (Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press) pp. 31–70
Morley SJ, Curtis PS and Pain VM . (1997) eIF4G: Translation's mystery factor begins to yield its secrets. RNA Publ. RNA Soc. 3: 1085–1104
Hentze M . (1997) eIF4G: A multipurpose ribosome adaptor? Science 275: 500–501
Gradi A, Imataka H, Svitkin YV, Rom E, Raught B, Morino S and Sonenberg N . (1998) A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18: 334–342
Yan R, Rychlik W, Etchison D and Rhoads RE . (1992) Amino acid sequence of the human protein synthesis initiation factor eIF-4gamma. J. Biol. Chem. 267: 23226–23231
Piron M, Vende P, Cohen J and Poncet D . (1998) Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 17: 5811–5821
Imataka H, Gradi A and Sonenberg N . (1998) A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17: 7480–7489
Johannes G and Sarnow P . (1998) Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA Publ. RNA Soc. 4: 1500–1513
Merrick WC . (1994) Eukaryotic protein synethesis: An in vitro analysis. Biochimie 76: 822–830
Mader S, Lee H, Pause A and Sonenberg N . (1995) The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 15: 4990–4997
Lamphear BJ, Kirchweger R, Skern T and Rhoads RE . (1995) Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases – implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270: 21975–21983
Tarun Jr SZ and Sachs AB . (1996) Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15: 7168–7177
Le H, Tanguay RL, Balasta ML, Wei CC, Browning KS, Metz AM, Goss DJ and Gallie DR . (1997) Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J. Biol. Chem. 272: 16247–16255
Tarun Jr SZ, Wells SE, Deardorff JA and Sachs AB . (1997) Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl. Acad. Sci. USA 94: 9046–9051
Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T and Sonenberg N . (1999) Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J. 18: 270–279
Haghighat A and Sonenberg N . (1997) eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 272: 21677–21680
Morley SJ and McKendrick L . (1997) Involvement of stress-activated protein kinase and p38/RK mitogen-activated protein kinase signaling pathways in the enhanced phosphorylation of initiation factor 4E in NIH 3T9 cells. J. Biol. Chem. 272: 17887–17893
Flynn A and Proud CG . (1996) Insulin-stimulated phosphorylation of initiation factor 4E is mediated by the MAP kinase pathway. FEBS Lett. 389: 162–166
Mendez R, Myers Jr MG, White MF and Rhoads RE . (1996) Stimulation of protein synthesis, eukaryotic translation initiation factor 4Ephosphorylation, and PHAS-I phosphorylation by insulin requires insulin receptor substrate 1 and phosphatidylinositol 3-kinase. Mol. Cell. Biol. 16: 2857–2864
Sonenberg N . (1996) mRNA 5′ cap-binding protein eIF4E and control of cell growth. In Translational Control, Hershey JWB, Mathews MB and Sonenberg N, eds (Cold Spring Harbor: Cold Spring Harbor Laboratory Press) pp. 245–269
Lazaris-Karatzas A, Montine KS and Sonenberg N . (1990) Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345: 544–547
Fukuchi-Shimogori T, Ishii I, Kashiwagi K, Mashiba H, Ekimoto H and Igarashi K . (1997) Malignant transformation by overproduction of translation initiation factor eIF4G. Cancer Res. 57: 5041–5044
Brass N, Heckel D, Sahin U, Pfreundschuh M, Sybrecht GW and Meese E . (1997) Translation initiation factor eIF-4gamma is encoded by an amplified gene and induces an immune response in squamous cell lung carcinoma. Hum. Mol. Genet. 6: 33–39
Nathan CAO, Liu L, Li BD, Abreo FW, Nandy I and De Benedetti A . (1997) Detection of the proto-oncogene eIF4E in surgical margins may predict recurrence in head and neck cancer. Oncogene 15: 579–584
Nathan CA, Carter P, Liu L, Li BD, Abreo F, Tudor A, Zimmer SG and De Benedetti A . (1997) Elevated expression of eIT4E and FGF-2 isoforms during vascularization of breast carcinomas. Oncogene 15: 1087–1094
Svitkin YV, Gradi A, Imataka H, Morino S and Sonenberg N . (1999) Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with initiation of host cell protein synthesis after human rhinovirus infection. J. Virol. 73: 3467–3472
Lamphear BJ, Yan R, Yang F, Waters D, Liebig H-D, Klump H, Kuechler E, Skern T and Rhoads RE . (1993) Mapping the cleavage site in protein synthesis initiation factor eIF-4gamma of the 2A proteases from human Coxsackievirus and rhinovirus. J. Biol. Chem. 268: 19200–19203
Ohlmann T, Rau M, Pain VM and Morley SJ . (1996) The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 15: 1371–1382
Ziegler E, Borman AM, Kirchweger R, Skern T and Kean KM . (1995) Foot-and-mouth disease virus Lb proteinase can stimulate rhinovirus and enterovirus IRES-driven translation and cleave several proteins of cellular and viral origin. J. Virol. 69: 3465–3474
Ziegler E, Borman AM, Deliat FG, Liebig HD, Jugovic D, Kean KM, Skern T and Kuechler E . (1995) Picornavirus 2A proteinase-mediated stimulation of internal initiation of translation is dependent on enzymatic activity and the cleavage products of cellular proteins. Virology 213: 549–557
Borman AM, Kirchweger R, Ziegler E, Rhoads RE, Skern T and Kean KM . (1997) eIF4G and its proteolytic cleavage products: Effect on initiation of protein syn-thesis from capped, uncapped, and IRES-containing mRNAs. RNA 3: 186–196
De Gregorio E, Preiss T and Hentze MW . (1998) Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end-dependent. RNA Publ. RNA Soc. 4: 828–836
Pestova TV, Shatsky IN and Hellen CUT . (1996) Functional dissection of eukaryotic initiation factor 4F: The 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 16: 6870–6878
Clemens MJ, Bushell M and Morley SJ . (1998) Degradation of eukaryotic polypeptide chain initiation factor (eIF) 4G in response to induction of apoptosis in human lymphoma cell lines. Oncogene 17: 2921–2931
Morley SJ, McKendrick L and Bushell M . (1998) Cleavage of translation initiation factor 4G (eIF4G) during anti-Fas IgM-induced apoptosis does not require signalling through the p38 mitogen-activated protein (MAP) kinase. FEBS Lett. 438: 41–48
Marissen WE and Lloyd RE . (1998) Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol. Cell. Biol. 18: 7565–7574
Bushell M, McKendrick L, Janicke RU, Clemens MJ and Morley SJ . (1999) Caspase-3 is necessary and sufficient for cleavage of protein synthesis eukaryotic initiation factor 4G during apoptosis. FEBS Lett. 451: 332–336
Bushell M, Wood W, Clemens MJ and Morley SJ . (2000) Changes in integrity and association of eukaryotic protein synthesis initiation factors during apoptosis. Eur. J. Biochem. 267: 1083–1091
Fraser CS, Pain VM and Morley SJ . (1999) The association of initiation factor 4F with Poly(A)-binding protein is enhanced in serum-stimulated Xenopus kidney cells. J. Biol. Chem. 274: 196–204
Rau M, Ohlmann T, Morley SJ and Pain VM . (1996) A reevaluation of the cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J. Biol. Chem. 271: 8983–8990
Ohlmann T, Pain VM, Wood W, Rau M and Morley SJ . (1997) The proteolytic cleavage of eukaryotic initiation factor (eIF) 4G is prevented by eIF4E binding protein (PHAS-I;4E-BP1) in the reticulocyte lysate. EMBO J. 16: 844–855
Clemens MJ, Bushell M, Jeffrey IW, Pain VM and Morley SJ . (2000) Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ. 7: 603–615
Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D, Ghayur T, Brady KD and Wong WW . (1997) Substrate specificities of caspase family proteases. J. Biol. Chem. 272: 9677–9682
Imataka H and Sonenberg N . (1997) Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 17: 6940–6947
De Gregorio E, Preiss T and Hentze M . (1999) Translation driven by an eIF4G core domain in vivo. EMBO J. 18: 4865–4874
Morino S, Imataka H, Svitkin YV, Pestova TV and Sonenberg N . (2000) Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 20: 468–477
Imataka H, Olsen HS and Sonenberg N . (1997) A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 16: 817–825
Levy-Strumpf N, Deiss LP, Berissi H and Kimchi A . (1997) DAP-5, a novel homolog of eukaryotic translation initiation factor 4G isolated as a putative modulator of gamma interferon-induced programmed cell death. Mol. Cell. Biol. 17: 1615–1625
Yamanaka S, Poksay KS, Arnold KS and Innerarity TL . (1997) A novel trans-lational repressor mRNA is edited in livers containing tumors caused by the trans-gene expression of the ApoB-mRNA editing enzyme. Genes Dev. 11: 321–333
Morley SJ and Pain VM . (1995) Hormone-induced meiotic maturation in Xenopus oocytes occurs independently of p70s6k activation and is associated with enhanced initiation factor (eIF)-4F phosphorylation and complex formation. J. Cell Sci. 108: 1751–1760
Morley SJ and Pain VM . (1995) Translational regulation during activation of porcine peripheral blood lymphocytes: Association and phosphorylation of the α and gamma subunits of the initiation factor complex eIF-4F. Biochem. J. 312: 627–635
Lloyd RE, Jense HG and Ehrenfeld E . (1987) Restriction of translation of capped mRNA in vitro as a model for poliovirus-induced inhibition of host-cell protein synthesis – relationship to p200 cleavage. J. Virol. 61: 2480–2488
Acknowledgements
We thank Dr Anne Gatignol (ICGM, Paris) for the gift of the Hela cell cDNA library, P Vende (Jouy-en-Josas, France) for the cloning and sequencing of eIF4GI constructs, Dr R Rhoads (Shreveport, Louisiana, USA) for the gift of pHFC1, A Gillman-Smith for help with mass spectrometry and N Castagné for skilful technical assistance. We also thank Dr Jenny Pain, Dr Linda McKendrick, Dr Ruth Simon, Ms Wendy Wood and Ms Gill Carpenter for helpful discussions. This research was supported by grants from The Royal Society, The Leukaemia Research Fund, The Cancer Prevention Research Trust and The Wellcome Trust (grant numbers 045619 and 056778). During the performance of this work M Bushell was funded by a BBSRC Industrial CASE Studentship in SJ Morley's laboratory, in collaboration with Roche Discovery (Welwyn Garden City). SJ Morley is a Senior Research Fellow of the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by RA Knight
Rights and permissions
About this article
Cite this article
Bushell, M., Poncet, D., Marissen, W. et al. Cleavage of polypeptide chain initiation factor eIF4GI during apoptosis in lymphoma cells: characterisation of an internal fragment generated by caspase-3-mediated cleavage. Cell Death Differ 7, 628–636 (2000). https://doi.org/10.1038/sj.cdd.4400699
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4400699
Keywords
This article is cited by
-
Heterogeneity and specialized functions of translation machinery: from genes to organisms
Nature Reviews Genetics (2018)
-
A researcher’s guide to the galaxy of IRESs
Cellular and Molecular Life Sciences (2017)
-
A novel mechanism for Bcr-Abl action: Bcr-Abl-mediated induction of the eIF4F translation initiation complex and mRNA translation
Oncogene (2007)
-
Inhibition of Cap-initiation complexes linked to a novel mechanism of eIF4G depletion in acute myocardial ischemia
Cell Death & Differentiation (2006)
-
Direct ribosomal binding by a cellular inhibitor of translation
Nature Structural & Molecular Biology (2006)