Abstract
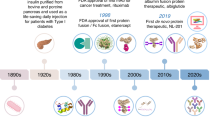
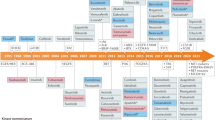
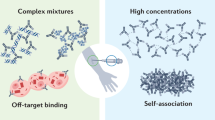
The modern biopharmaceutical industry traces its roots to the dawn of the twentieth century, coincident with marketing of aspirin—a signature event in the history of modern drug development. Although the archetypal discovery process did not change markedly in the first seven decades of the industry, the past fifty years have seen two successive waves of transformative innovation in the development of drug molecules: the rise of ‘rational drug discovery’ methodology in the 1970s, followed by the invention of recombinant protein-based therapeutic agents in the 1980s. An incipient fourth wave is the advent of multispecific drugs. The successful development of prospectively designed multispecific drugs has the potential to reconfigure our ideas of how target-based therapeutic molecules can work, and what it is possible to achieve with them. Here I review the two major classes of multispecific drugs: those that enrich a therapeutic agent at a particular site of action and those that link a therapeutic target to a biological effector. The latter class—being freed from the constraint of having to directly modulate the target upon binding—may enable access to components of the proteome that currently cannot be targeted by drugs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Madsen, U., Krogsgaard-Larsen, P. & Liljefors, T. Textbook of Drug Design and Discovery (Taylor & Francis, 2002).
Drews, J. Drug discovery: a historical perspective. Science 287, 1960–1964 (2000).
Patlak, M. From viper’s venom to drug design: treating hypertension. FASEB J. 18, 421 (2004).
Altman, L. K. A new insulin given approval for use in U.S. NY Times (30 October 1982).
Ortho Multicenter Transplant Study Group. A randomized clinical trial of OKT3 monoclonal antibody for acute rejection of cadaveric renal transplants. N. Engl. J. Med. 313, 337–342 (1985).
Labrijn, A. F., Janmaat, M. L., Reichert, J. M. & Parren, P. W. H. I. Bispecific antibodies: a mechanistic review of the pipeline. Nat. Rev. Drug Discov. 18, 585–608 (2019).
Nair, J. K. et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 136, 16958–16961 (2014).A key paper that established the utility of using N-acetylglucosamine to specify liver uptake of siRNAs.
Springer, A. D. & Dowdy, S. F. GalNAc–siRNA conjugates: leading the way for delivery of RNAi therapeutics. Nucleic Acid Ther. 28, 109–118 (2018).
Alderson, R. F. et al. CAT-8015: a second-generation pseudomonas exotoxin A-based immunotherapy targeting CD22-expressing hematologic malignancies. Clin. Cancer Res. 15, 832–839 (2009).
Abdollahpour-Alitappeh, M. et al. Antibody–drug conjugates (ADCs) for cancer therapy: strategies, challenges, and successes. J. Cell. Physiol. 234, 5628–5642 (2019).
Beck, A., Goetsch, L., Dumontet, C. & Corvaïa, N. Strategies and challenges for the next generation of antibody–drug conjugates. Nat. Rev. Drug Discov. 16, 315–337 (2017).
Neri, D. Antibody–cytokine fusions: versatile products for the modulation of anticancer immunity. Cancer Immunol. Res. 7, 348–354 (2019).
Stanton, B. Z., Chory, E. J. & Crabtree, G. R. Chemically induced proximity in biology and medicine. Science 359, eaao5902 (2018).
Kolata, G. FDA speeds approval of cyclosporin. Science 221, 1273 (1983).
Liu, J. et al. Calcineurin is a common target of cyclophilin–cyclosporin A and FKBP–FK506 complexes. Cell 66, 807–815 (1991). A landmark paper that established an unexpected mode of drug action via neocomplex formation.
Maniaci, C. & Ciulli, A. Bifunctional chemical probes inducing protein–protein interactions. Curr. Opin. Chem. Biol. 52, 145–156 (2019).
Griffith, J. P. et al. X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12–FK506 complex. Cell 82, 507–522 (1995).
Brown, E. J. et al. A mammalian protein targeted by G1-arresting rapamycin–receptor complex. Nature 369, 756–758 (1994).
Gray, W. M., Kepinski, S., Rouse, D., Leyser, O. & Estelle, M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276 (2001).
Tan, X. et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645 (2007). Auxin works by a remarkable mechanism, whereby it binds at the interface between ubiquitin ligase subunit TIR1 and a substrate protein, stabilizing the ternary complex and inducing ubiquitination and degradation of the substrate protein.
Fujiwara, T., Oda, K., Yokota, S., Takatsuki, A. & Ikehara, Y. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J. Biol. Chem. 263, 18545–18552 (1988).
Lippincott-Schwartz, J., Yuan, L. C., Bonifacino, J. S. & Klausner, R. D. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56, 801–813 (1989).
Peyroche, A. et al. Brefeldin A acts to stabilize an abortive ARF–GDP–Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3, 275–285 (1999).
Mossessova, E., Corpina, R. A. & Goldberg, J. Crystal structure of ARF1*Sec7 complexed with brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol. Cell 12, 1403–1411 (2003).These two papers 23,24 establish that brefeldin A sits at the interface between ARF and SEC7 to stabilize what is normally a very dynamic protein–protein interaction, pointing to a novel mechanism for drug action.
Chardin, P. & McCormick, F. Brefeldin A: the advantage of being uncompetitive. Cell 97, 153–155 (1999).
Palacino, J. et al. SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat. Chem. Biol. 11, 511–517 (2015).
Gandhi, A. K. et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN.). Br. J. Haematol. 164, 811–821 (2014).
Krönke, J. et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305 (2014).
Lu, G. et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309 (2014). These three papers 27–29report the discovery of a suprising mechanism of action for one of the best-selling drugs in the world; similar to auxin, lenalidomide stabilizes an interaction between a substrate (IKZF1 and/or IKZF3) and an ubiquitin ligase (CRBN), resulting in ubiquitination and degradation of the IKZF proteins.
Speirs, A. L. Thalidomide and congenital abnormalities. Lancet 279, 303–305 (1962).
Sheskin, J. Thalidomide in the treatment of lepra reactions. Clin. Pharmacol. Ther. 6, 303–306 (1965).
Larkin, M. Low-dose thalidomide seems to be effective in multiple myeloma. Lancet 354, 925 (1999).
Muller, G. W. et al. Amino-substituted thalidomide analogs: potent inhibitors of TNF-alpha production. Bioorg. Med. Chem. Lett. 9, 1625–1630 (1999).
Ito, T. et al. Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350 (2010). This seminal paper set in motion the series of discoveries that culminated in the unravelling of the unusual mechanism of action of lenalidomide.
Lopez-Girona, A. et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia 26, 2326–2335 (2012).
Zhu, Y. X., Kortuem, K. M. & Stewart, A. K. Molecular mechanism of action of immune-modulatory drugs thalidomide, lenalidomide and pomalidomide in multiple myeloma. Leuk. Lymphoma 54, 683–687 (2013).
Cortés, M. & Georgopoulos, K. Aiolos is required for the generation of high affinity bone marrow plasma cells responsible for long-term immunity. J. Exp. Med. 199, 209–219 (2004).
Han, T. et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 356, eaal3755 (2017).
Uehara, T. et al. Selective degradation of splicing factor CAPERα by anticancer sulfonamides. Nat. Chem. Biol. 13, 675–680 (2017).
Sakamoto, K. M. et al. Protacs: chimeric molecules that target proteins to the Skp1–Cullin-F box complex for ubiquitination and degradation. Proc. Natl Acad. Sci. USA 98, 8554–8559 (2001). This work provided the first demonstration that it is possible to specify the degradation of a defined target by inducing its proximity to an ubiquitin ligase using a synthetic molecule.
Sakamoto, K. M. et al. Development of protacs to target cancer-promoting proteins for ubiquitination and degradation. Mol. Cell. Proteomics 2, 1350–1358 (2003).
Schneekloth, J. S. Jr et al. Chemical genetic control of protein levels: selective in vivo targeted degradation. J. Am. Chem. Soc. 126, 3748–3754 (2004).
Pettersson, M. & Crews, C. M. Proteolysis targeting chimeras (PROTACs) - past, present and future. Drug Discov. Today. Technol. 31, 15–27 (2019).
Verma, R., Mohl, D. & Deshaies, R. J. Harnessing the power of proteolysis for targeted protein inactivation. Mol. Cell 77, 446–460 (2020).
Buckley, D. L. et al. Small-molecule inhibitors of the interaction between the E3 ligase VHL and HIF1α. Angew. Chem. Int. Edn Engl. 51, 11463–11467 (2012).
Bondeson, D. P. et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 11, 611–617 (2015).
Winter, G. E. et al. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381 (2015).
Zengerle, M., Chan, K. H. & Ciulli, A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem. Biol. 10, 1770–1777 (2015).
Lu, J. et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem. Biol. 22, 755–763 (2015).These four papers 46–49established that it is possible to construct highly potent and selective Protacs that work in vivo using ligands that bind ubiquitin ligases VHL or CRBN.
Arvinas, Inc. Clinical trial of ARV-471 in patients with ER+/HER2- locally advanced or metastatic breast cancer (mBC). https://clinicaltrials.gov/ct2/show/NCT04072952 (2019).
Flanagan, J. J. & Neklesa, T. K. Targeting nuclear receptors with PROTAC degraders. Mol. Cell. Endocrinol. 493, 110452 (2019).
Salami, J. et al. Androgen receptor degradation by the proteolysis-targeting chimera ARCC-4 outperforms enzalutamide in cellular models of prostate cancer drug resistance. Commun. Biol. 1, 100 (2018).
Arvinas, Inc. A phase 1 clinical trial of ARV-110 in patients with metastatic castration-resistant prostate cancer (mCRPC). https://clinicaltrials.gov/ct2/show/NCT03888612 (2019).
Perez, P., Hoffman, R. W., Shaw, S., Bluestone, J. A. & Segal, D. M. Specific targeting of cytotoxic T cells by anti-T3 linked to anti-target cell antibody. Nature 316, 354–356 (1985).
Staerz, U. D. & Bevan, M. J. Hybrid hybridoma producing a bispecific monoclonal antibody that can focus effector T-cell activity. Proc. Natl Acad. Sci. USA 83, 1453–1457 (1986).These two papers 54,55launched the field of bispecific CD3 engagers for use in cancer immunotherapy.
Riechelmann, H. et al. Adoptive therapy of head and neck squamous cell carcinoma with antibody coated immune cells: a pilot clinical trial. Cancer Immunol. Immunother. 56, 1397–1406 (2007).
Löffler, A. et al. A recombinant bispecific single-chain antibody, CD19 x CD3, induces rapid and high lymphoma-directed cytotoxicity by unstimulated T lymphocytes. Blood 95, 2098–2103 (2000).
Bargou, R. et al. Tumor regression in cancer patients by very low doses of a T cell-engaging antibody. Science 321, 974–977 (2008). More than 20 years after research on bispecific CD3 engagers began, this paper provided the first compelling demonstration of the tremendous clinical potential of this class of bispecific molecules.
Curran, E. & Stock, W. Taking a “BiTE out of ALL”: blinatumomab approval for MRD-positive ALL. Blood 133, 1715–1719 (2019).
Przepiorka, D. et al. FDA approval: blinatumomab. Clin. Cancer Res. 21, 4035–4039 (2015).
Viardot, A. & Bargou, R. Bispecific antibodies in haematological malignancies. Cancer Treat. Rev. 65, 87–95 (2018).
Gauthier, L. et al. Multifunctional natural killer cell engagers targeting NKp46 trigger protective tumor immunity. Cell 177, 1701–1713 (2019).
Nayyar, G., Chu, Y. & Cairo, M. S. Overcoming resistance to natural killer cell based immunotherapies for solid tumors. Front. Oncol. 9, 51 (2019).
Sampei, Z. et al. Identification and multidimensional optimization of an asymmetric bispecific IgG antibody mimicking the function of factor VIII cofactor activity. PLoS ONE 8, e57479 (2013). This paper reports on the long campaign that led to discovery of emicizumab, a landmark achievement in the history of bispecific biologic drugs.
Kitazawa, T. & Shima, M. Emicizumab, a humanized bispecific antibody to coagulation factors IXa and X with a factor VIIIa-cofactor activity. Int. J. Hematol. 111, 20–30 (2018).
Foster, D. J. et al. Advanced siRNA designs further improve in vivo performance of GalNAc–siRNA conjugates. Mol. Ther. 26, 708–717 (2018).
Khvorova, A. & Watts, J. K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 35, 238–248 (2017).
Egli, M. & Manoharan, M. Re-engineering RNA molecules into therapeutic agents. Acc. Chem. Res. 52, 1036–1047 (2019).
Matsuda, S. et al. siRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem. Biol. 10, 1181–1187 (2015).
Levin, A. A. Treating disease at the RNA level with oligonucleotides. N. Engl. J. Med. 380, 57–70 (2019).
Winkler, J. Extrahepatic targeting of oligonucleotides with receptor-binding non-immunoglobulin scaffold proteins. Nucleic Acid Ther. 28, 137–145 (2018).
Matthews, P. M. Chronic inflammation in multiple sclerosis – seeing what was always there. Nat. Rev. Neurol. 15, 582–593 (2019).
Freskgård, P. O. & Urich, E. Antibody therapies in CNS diseases. Neuropharmacology 120, 38–55 (2017).
Poduslo, J. F., Curran, G. L. & Berg, C. T. Macromolecular permeability across the blood–nerve and blood–brain barriers. Proc. Natl Acad. Sci. USA 91, 5705–5709 (1994).
Sevigny, J. et al. The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease. Nature 537, 50–56 (2016).
Lajoie, J. M. & Shusta, E. V. Targeting receptor-mediated transport for delivery of biologics across the blood–brain barrier. Annu. Rev. Pharmacol. Toxicol. 55, 613–631 (2015).
Mäger, I. et al. Targeting blood–brain-barrier transcytosis – perspectives for drug delivery. Neuropharmacology 120, 4–7 (2017).
Pardridge, W. M. Delivery of biologics across the blood–brain barrier with molecular Trojan horse technology. BioDrugs 31, 503–519 (2017).
Zuchero, Y. J. et al. Discovery of novel blood–brain barrier targets to enhance brain uptake of therapeutic antibodies. Neuron 89, 70–82 (2016).
Stutz, C. C., Zhang, X. & Shusta, E. V. Combinatorial approaches for the identification of brain drug delivery targets. Curr. Pharm. Des. 20, 1564–1576 (2014).
Chan, K. Y. et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179 (2017).
Le, R. Q. et al. FDA approval summary: tocilizumab for treatment of chimeric antigen receptor T cell-induced severe or life-threatening cytokine release syndrome. Oncologist 23, 943–947 (2018).
Li, J. et al. CD3 bispecific antibody-induced cytokine release is dispensable for cytotoxic T cell activity. Sci. Transl. Med. 11, eaax8861 (2019). These authors report that it is possible to uncouple cytokine release from the anti-tumour activity of BCEs, which could lead to an enhanced therapeutic index for BCEs.
Topp, M. S. et al. Long-term follow-up of hematologic relapse-free survival in a phase 2 study of blinatumomab in patients with MRD in B-lineage ALL. Blood 120, 5185–5187 (2012).
Balakrishnan, A. et al. Multispecific targeting with synthetic ankyrin repeat motif chimeric antigen receptors. Clin. Cancer Res. 25, 7506–7516 (2019).
Zhao, J., Song, Y. & Liu, D. Clinical trials of dual-target CAR T cells, donor-derived CAR T cells, and universal CAR T cells for acute lymphoid leukemia. J. Hematol. Oncol. 12, 17 (2019).
Kebenko, M. et al. A multicenter phase 1 study of solitomab (MT110, AMG 110), a bispecific EpCAM/CD3 T-cell engager (BiTE®) antibody construct, in patients with refractory solid tumors. OncoImmunology 7, e1450710 (2018).
Mau-Sørensen, M. et al. A phase I trial of intravenous catumaxomab: a bispecific monoclonal antibody targeting EpCAM and the T cell coreceptor CD3. Cancer Chemother. Pharmacol. 75, 1065–1073 (2015).
Desnoyers, L. R. et al. Tumor-specific activation of an EGFR-targeting probody enhances therapeutic index. Sci. Transl. Med. 5, 207ra144 (2013).
Kloss, C. C., Condomines, M., Cartellieri, M., Bachmann, M. & Sadelain, M. Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat. Biotechnol. 31, 71–75 (2013).
Roybal, K. T. et al. Precision tumor recognition by T cells with combinatorial antigen-sensing circuits. Cell 164, 770–779 (2016).
Srivastava, S. et al. Logic-gated ROR1 chimeric antigen receptor expression rescues T cell-mediated toxicity to normal tissues and enables selective tumor targeting. Cancer Cell 35, 489–503 (2019).
Langan, R. A. et al. De novo design of bioactive protein switches. Nature 572, 205–210 (2019) Computational protein design is used to construct a synthetic AND gate that may have applicability to future therapeutic agents.
de Sostoa, J. et al. Targeting the tumor stroma with an oncolytic adenovirus secreting a fibroblast activation protein-targeted bispecific T-cell engager. J. Immunother. Cancer 7, 19 (2019).
Brey, C. U. et al. A gB/CD3 bispecific BiTE antibody construct for targeting human cytomegalovirus-infected cells. Sci. Rep. 8, 17453 (2018).
Brozy, J. et al. Antiviral activity of HIV gp120-targeting bispecific T cell engager antibody constructs. J. Virol. 92, e00491-18 (2018).
Aghajanian, H. et al. Targeting cardiac fibrosis with engineered T cells. Nature 573, 430–433 (2019).
Ellebrecht, C. T. et al. Reengineering chimeric antigen receptor T cells for targeted therapy of autoimmune disease. Science 353, 179–184 (2016).
Baker, D. J. et al. Naturally occurring p16Ink4a-positive cells shorten healthy lifespan. Nature 530, 184–189 (2016).
Igawa, T. et al. Engineered monoclonal antibody with novel antigen-sweeping activity in vivo. PLoS ONE 8, e63236 (2013).
Banik, S., Pedram, K., Wisnovsky, S., Riley, N. & Bertozzi, C. Lysosome targeting chimeras (LYTACs) for the degradation of secreted and membrane proteins. Preprint at https://chemrxiv.org/articles/Lysosome_Targeting_Chimeras_LYTACs_for_the_Degradation_of_Secreted_and_Membrane_Proteins/7927061 (2019).
Takahashi, D. et al. AUTACs: cargo-specific degraders using selective autophagy. Mol. Cell 76, 797–810.e10 (2019).
Ito, C. et al. Endogenous nitrated nucleotide is a key mediator of autophagy and innate defense against bacteria. Mol. Cell 52, 794–804 (2013
Yamazoe, S. et al. Heterobifunctional molecules induce dephosphorylation of kinases-a proof of concept study. J. Med. Chem. https://doi.org/10.1021/acs.jmedchem.9b01167 (2020).
Zhang, Z. & Shokat, K. M. Bifunctional small-molecule ligands of K-Ras induce its association with immunophilin proteins. Angew. Chem. Int. Edn Engl. 58, 16314–16319 (2019).
Costales, M. G. et al. Small-molecule targeted recruitment of a nuclease to cleave an oncogenic RNA in a mouse model of metastatic cancer. Proc. Natl Acad. Sci. USA 117, 2406–2411 (2020). This study reports on ribonuclease-targeting chimaeras, which potentially represent an entire new class of therapeutic agent that targets the elimination of specific RNAs.
Klemm, J. D., Beals, C. R. & Crabtree, G. R. Rapid targeting of nuclear proteins to the cytoplasm. Curr. Biol. 7, 638–644 (1997).
Haruki, H., Nishikawa, J. & Laemmli, U. K. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell 31, 925–932 (2008).
Wang, Q. et al. Design and production of bispecific antibodies. Antibodies (Basel) 8, 43 (2019).
Bratt, J., Linderholm, A., Monroe, B. & Chamow, S. Therapeutic IgG-like bispecific antibodies. Bioprocess Int. 16, 40–49 (2018).
Ponce, R. et al. Immunogenicity of biologically-derived therapeutics: assessment and interpretation of nonclinical safety studies. Regul. Toxicol. Pharmacol. 54, 164–182 (2009).
Datta-Mannan, A. et al. Aberrant bispecific antibody pharmacokinetics linked to liver sinusoidal endothelium clearance mechanism in cynomolgus monkeys. MAbs 8, 969–982 (2016).
Harper, J. et al. An approved in vitro approach to preclinical safety and efficacy evaluation of engineered T cell receptor anti-CD3 bispecific (ImmTAC) molecules. PLoS ONE 13, e0205491 (2018).
Saber, H., Del Valle, P., Ricks, T. K. & Leighton, J. K. An FDA oncology analysis of CD3 bispecific constructs and first-in-human dose selection. Regul. Toxicol. Pharmacol. 90, 144–152 (2017).
US Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research & Center for Biologics Evaluation and Research. Bispecific antibody development programs Guidance for industry, https://www.fda.gov/media/123313/download (2019).
Lipinski, C. A., Lombardo, F., Dominy, B. W. & Feeney, P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 23, 3–25 (1997).
Dauvois, S., Danielian, P. S., White, R. & Parker, M. G. Antiestrogen ICI 164,384 reduces cellular estrogen receptor content by increasing its turnover. Proc. Natl Acad. Sci. USA 89, 4037–4041 (1992).
Bernstein, I. D. Monoclonal antibodies to the myeloid stem cells: therapeutic implications of CMA-676, a humanized anti-CD33 antibody calicheamicin conjugate. Leukemia 14, 474–475 (2000).
Guan, J. et al. Therapeutic ligands antagonize estrogen receptor function by impairing its mobility. Cell 178, 949–963 (2019).
Du, J. et al. Structural basis for the blockage of IL-2 signaling by therapeutic antibody basiliximab. J. Immunol. 184, 1361–1368 (2010).
Gadd, M. S. et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 13, 514–521 (2017).
Acknowledgements
I thank C. Kiefer and L. Jia for artwork, J. Bailis, R. Desphande, H. Hsu, G. Moffat, J. Murry, N. Ogbechie, and D. Rock for supplying content and comments, and D. Baker, M. Chu-Moyer, A. Gouliaev, S. Haldar, E. Levy, F. Martin, D. Reese, E. Sigal, K. Stefánsson, P. Tagari, and S. Wang for comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
R.J.D. is an employee, officer and shareholder of Amgen.
Additional information
Peer review information Nature thanks Alessio Ciulli, Ira Mellman and Nurulain Zaveri for their contribution to the peer review of this work.
Supplementary information
Supplementary Tables
This file contains Supplementary Tables 1-3.
Rights and permissions
About this article
Cite this article
Deshaies, R.J. Multispecific drugs herald a new era of biopharmaceutical innovation. Nature 580, 329–338 (2020). https://doi.org/10.1038/s41586-020-2168-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2168-1
This article is cited by
-
Proteome-scale discovery of protein degradation and stabilization effectors
Nature (2024)
-
Beyond canonical PROTAC: biological targeted protein degradation (bioTPD)
Biomaterials Research (2023)
-
M2 macrophage-derived cathepsin S promotes peripheral nerve regeneration via fibroblast–Schwann cell-signaling relay
Journal of Neuroinflammation (2023)
-
Affinity and cooperativity modulate ternary complex formation to drive targeted protein degradation
Nature Communications (2023)
-
AI can help to speed up drug discovery — but only if we give it the right data
Nature (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.