Abstract
Purpose
We investigated the frequencies and characteristics of intragenic copy-number variants (CNVs) in a deep sampling of disease genes associated with monogenic disorders.
Methods
Subsets of 1507 genes were tested using next-generation sequencing to simultaneously detect sequence variants and CNVs in >143,000 individuals referred for genetic testing. We analyzed CNVs in gene panels for hereditary cancer syndromes and cardiovascular, neurological, or pediatric disorders.
Results
Our analysis identified 2844 intragenic CNVs in 384 clinically tested genes. CNVs were observed in 1.9% of the entire cohort but in a disproportionately high fraction (9.8%) of individuals with a clinically significant result. CNVs accounted for 4.7–35% of pathogenic variants, depending on clinical specialty. Distinct patterns existed among CNVs in terms of copy number, location, exons affected, clinical classification, and genes affected. Separately, analysis of de-identified data for 599 genes unrelated to the clinical phenotype yielded 4054 CNVs. Most of these CNVs were novel rare events, present as duplications, and enriched in genes associated with recessive disorders or lacking loss-of-function mutational mechanisms.
Conclusion
Universal intragenic CNV analysis adds substantial clinical sensitivity to genetic testing. Clinically relevant CNVs have distinct properties that distinguish them from CNVs contributing to normal variation in human disease genes.
Similar content being viewed by others
Introduction
Within the broad spectrum of variation in our genome, relatively little is known about the prevalence of intragenic deletions and duplications, or copy-number variants (CNVs), beyond those responsible for a small number of hereditary disorders such as Duchenne muscular dystrophy and BRCA1-associated breast cancer. Exon-level copy-number detection methods are used in genetic testing for these disorders but are not universally applied to other genes, which greatly limits our knowledge of intragenic CNV prevalence across the medically relevant portions of the genome. Although exome-based approaches are beginning to shed light on these CNVs, the resolution and confidence of CNV calling with these methods remains limited.1,2 Exon-focused microarray-based comparative genomic hybridization (array CGH) provides high resolution for CNV analysis but is not used universally.3,4,5 Array CGH is now being replaced by highly sensitive and scalable targeted next-generation sequencing (NGS) methods, providing an opportunity to investigate CNVs broadly across disease genes.6,7,8,9
CNVs also explain a proportion of the benign variation in the human genome. Microarray and NGS methods are illuminating this variation in healthy individuals, as demonstrated by the 1000 Genomes Project, Exome Aggregation Consortium (ExAC) projects, and other large-scale efforts (Maxwell E, 2017, unpublished data).10,11,10,12 The data from these studies are being compiled in archives such as the Database of Genomic Variants (DGV) and ClinVar and have contributed to our understanding of dosage-sensitive regions of the genome.13,14 Investigating intragenic CNVs can further refine the resolution of this dosage map and complement our understanding of larger chromosomal CNVs.
We have developed and validated an NGS assay and bioinformatics pipeline to simultaneously identify sequence variants and intragenic CNVs with high sensitivity.9 Herein, we describe CNVs identified in a cohort of >143,000 unrelated individuals referred for diagnostic genetic testing for a suspected Mendelian disorder. In addition, we studied CNVs in disease genes unrelated to these patients’ phenotypes to estimate a baseline prevalence. Our data show that a substantial number of pathogenic variants are in fact intragenic CNVs that have not been routinely investigated across a wide range of disease genes in traditional genetic testing. We also show that clinically relevant CNVs are characteristically different from CNVs that are present as naturally existing variation in the genome.
Materials and methods
NGS assay and bioinformatics
Each gene on Invitae’s NGS panels was targeted with oligonucleotide baits (Agilent Technologies, Santa Clara, CA; Roche, Pleasanton, CA; IDT, Coralville, IA) to capture exons, the 10–20 bases flanking intronic sequences, and certain noncoding regions of clinical interest. Baits were iteratively balanced to obtain a minimum of 50× and an average of 350× depth-of-sequence read coverage. Invitae uses NGS assays (not exome–based) containing hundreds of genes that constitute many panels and designed to address differential diagnoses within clinical specialties. Our bioinformatics pipeline incorporates both community standard and custom algorithms to identify single-nucleotide variants (SNVs) and small indels, large indels, structural variants with breakpoints in target sequences, and exon-level CNVs.9,15
Patient samples and analysis
Blood, saliva, or genomic DNA from patients who were consented for diagnostic testing were processed by NGS as described previously.9,15 For this study, clinically reported CNVs were collected from Invitae’s variant database. In addition, data from genes unrelated to the presenting clinical phenotype were de-identified for analysis of baseline CNVs under institutional review board (IRB) approval (Western IRB, #20161796). A Pearson’s chi-squared test was used to compute p values for differences in observed CNV counts. Ninety-five percent confidence intervals (CIs) were calculated using the Wilson method.
Clinical classification of CNVs
The clinical interpretation of CNVs was based on their predicted effects on the transcript and established mutational mechanism for each gene. CNVs were classified using the five-tier system for grading evidence for pathogenicity as recommended by the American College of Medical Genetics and Genomics (ACMG).16 To ensure consistent evidence-based CNV interpretation and maintain congruence with ACMG’s sequence variant interpretation guidelines, we developed a point-based scoring system that includes criteria for classifying CNVs.17
Orthogonal confirmation
For clinically tested genes, we used array CGH to confirm CNVs if they had been classified as likely pathogenic or pathogenic (LP/P) or were single-exon CNVs classified as variants of uncertain significance (VUS). Multiplex ligation-dependent amplification (MRC Holland, Amsterdam, Netherlands) and long-range polymerase chain reaction (PCR) with Sanger sequencing were used to confirm CNVs in exons 12–15 of PMS2, which has a pseudogene copy. Deletions in SMN1 were confirmed with ligation-dependent sequencing, an NGS adaptation of multiplex ligation-dependent amplification. Baseline CNVs were not confirmed with array CGH if they had high NGS quality scores (Q99) or lower quality scores (Q35–Q98) but included multiple exons, because these types of CNVs always confirmed as true events in clinically tested genes. However, we excluded single-exon baseline CNVs flagged as low-quality calls.
Results
Prevalence of intragenic CNVs in a large clinical cohort
We tested various subsets of 1507 genes in 143,515 unrelated individuals referred for diagnostic NGS gene panel testing. A total of ~4.8 million single-gene analyses were completed. Among nearly 8.1 million variants of all types, we identified 2844 intragenic CNVs (1237 distinct events). These CNVs accounted for 0.03% of all variants, 3.1% of reported variants, and notably, 9.1% of variants classified as LP/P (Supplementary Table 1 and Supplementary Figure 1). These variants were found across 384 genes and included 1810 deletions and 1034 duplications, which together represented a prevalence of 1.9% in this cohort, 4.4% among individuals with at least one reported variant, and more significantly, 9.8% among individuals who received a report with an LP/P variant of any type.
Patterns of intragenic CNV occurrence
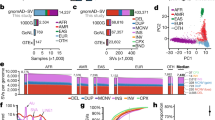
CNVs fell into one of three categories—single rare events, common recurrent events, and low-frequency recurrent events (Fig. 1a). Each category represented approximately one-third of all observed CNVs. The vast majority of the 384 genes with CNVs had just one CNV each, but these single CNVs together accounted for less than 10% of all events (Fig. 1b). By contrast, 31 of the 384 genes had 15 or more CNVs, but these represented nearly 70% of all CNVs. Aside from frequencies, the intragenic locations and sizes of CNVs were examined, because these properties can determine clinical impact. One-fourth of the CNVs included just one exon. A majority of intragenic CNVs were multi-exonic partial-gene events, and most encompassed only internal exons without involving the terminal (first or last) coding exons (Fig. 1c,d). Among partial-gene CNVs involving terminal exons, more deletions than duplications included the first exons, whereas a similar number of deletions and duplications included the last exons. Finally, a greater proportion of duplications than deletions included the full gene. Nearly one-fifth of all distinct (non-redundant) CNVs included a full gene, and in 40 instances, the CNVs encompassed several neighboring genes and were present on at least 10 chromosomes (Supplementary Tables 1, 2).
a Histogram showing the number of distinct CNVs observed in the tested genes. The columns in the chart indicate the number of times the CNVs were observed. The line graph shows the proportion of total observed CNVs in each frequency bin. For example, the first column shows that nearly 900 CNVs occurred just once and, in aggregate, accounted for roughly 30% of all CNVs. b Histogram showing the number of genes that contained CNVs in our clinical cohort. The columns in the chart show incremental increases in the number of CNVs observed in a gene. The line graph shows the proportion of CNVs at arbitrary increments of CNV occurrence per gene. For example, nearly 200 genes had just 1 CNV, which together accounted for less than 10% of all events. By contrast, approximately 30 genes had more than 15 CNVs each, which represented nearly 70% of all CNVs. c Distribution of deletions and duplications by number of exons affected and by clinical interpretation. Cytogenetic events are defined as contiguous CNVs of the same zygosity affecting neighboring genes on a single chromosome. Some whole-gene events may in fact be part of larger cytogenetic events but are not listed as such because other genes within the predicted cytogenetic event were absent from our assay and therefore unavailable for analysis. d Count of CNV duplications and deletions detected in clinical and baseline CNV data. CNVs are split into those including a whole gene (classes I, V), at least the last exon (classes II, VI), at least the first exon (classes III, VII), or only an internal exon(s) (classes IV, VIII). A generic gene structure is shown at the top. Green and purple boxes denote “terminal exons” and all others are “internal exons,” as described in the text. Empty boxes indicate deleted exons. This figure assumes that intragenic duplications occur in tandem, which is often the case with such events. CNVs involving just promoter regions are not represented in this figure. A Pearson’s chi-squared contingency table gives a p value of p < 1×10−5 for duplications and p = 1.5×10−5 for deletions, indicating that the difference in the distribution of CNVs across the gene is not merely due to sampling differences between clinical and baseline CNVs. e and f Deletions and duplications in clinically tested genes and their interpretations. The chart in (e) shows genes with loss-of-function (LOF) mutational mechanisms, and that in (f) shows genes without loss-of-function (LOF) mechanisms. Most genes included in our panels were curated as having LOF mechanisms. The clinical classification of each CNV, inheritance pattern of the gene with the CNV, and zygosity of the variants are compared. For X-linked (XL) genes, heterozygous CNVs in females are shown separately from CNVs in males. AD autosomal dominant, AR autosomal recessive, F female, het heterozygous, hom homozygous, M male, LP likely pathogenic variant, P pathogenic variant, VUS variants of uncertain significance. The “Pathogenic” label in c, e, and f includes CNVs classified as pathogenic and likely pathogenic
Clinical classification of CNVs
Deletions were more frequent in this clinical cohort, and most were reported as LP/P variants (Fig. 1c). However, a few deletions were classified as VUS, primarily because they were in-frame variants in genes without loss-of-function (LOF) mutational mechanisms. By contrast, more than half of the duplications were classified as VUS. Among partial-gene duplications, 359 involved terminal exons and 225 involved only internal exons (Fig. 1d). At least 166 duplications encompassing only internal exons were predicted to have an adverse effect on the transcript reading frame and therefore classified as LP/P (Supplementary Table 2). For at least 30 duplications, we observed putative breakpoints based on split-read sequence data and predicted a tandem arrangement that would disrupt the transcript reading frame. This supports previous assertions that intragenic duplications are typically localized tandem rearrangements versus more complicated events such as insertional translocations.18
We also considered the distribution and zygosity of CNVs in genes associated with autosomal dominant (AD), autosomal recessive (AR), and X-linked (XL) disorders (Fig. 1e,f). The vast majority of CNVs were in genes associated with AD or XL inheritance, although this outcome reflects a bias because most of the tested genes had these inheritance patterns. Of 2096 CNVs classified as LP/P, 85% were in genes associated with AD or XL inheritance and 15% were in genes associated with AR inheritance. Of the latter, 6.7% were homozygous deletions, 2.8% were compound heterozygous changes accompanying a pathogenic SNV on the other allele (constituting a positive molecular diagnosis for an AR disorder; Supplementary Table 1), and 5.5% were single heterozygous events.
Nearly all CNVs in this cohort were found in genes with LOF mechanisms (Fig. 1e). Most CNVs in these genes were deletions classified as pathogenic, whereas more than half of the duplications were classified as VUS. Comparatively, the 304 genes without LOF mechanisms had few CNVs, mostly classified as VUS or benign (Fig. 1f), and significantly more duplications than deletions (p = 1.8×10–9).
CNVs and morbidity
Analysis of a large number of multigene panels showed varying CNV prevalence across disease groups (Fig. 2a,b; Supplementary Table 4). Genes with CNVs had either mostly recurrent events, mostly unique events, or a mix of both (Fig. 2c). Among panels that had yielded at least 10 pathogenic variants of any type, more than one-third had CNVs accounting for greater than 10% of pathogenic variants. Gene panels yielding the highest number of CNVs were those for spinal muscular atrophy, Charcot–Marie–Tooth disease, and dystrophinopathies, as expected. However, panels for congenital heart defects and heterotaxy, Lynch syndrome, sarcoma, muscular dystrophy, and dystonia also identified many CNVs. By contrast, gene panels with the lowest CNV frequencies included those for chronic pancreatitis, RASopathies, cardiomyopathies, and hereditary thrombophilia.
a Panels in each clinical specialty are shown with the percentage of positive reports that included one or more pathogenic CNVs. Only panels that were clinically tested at least 100 times and had at least 10 pathogenic variants of any type are included. Each panel is represented by a circle. b Proportion of positive reports that included pathogenic CNVs in various panels. These panels were the top five with the most pathogenic variants in each clinical specialty. c Three patterns of CNV occurrence were discernible: recurrent events (e.g., in PMP22 and SMN1), predominantly rare unique events (e.g., in DMD and BRCA2), and a mix of both (e.g., in BRCA1 and MSH2). The number of distinct variants is shown in blue, and additional/recurrent observations of these variants are shown in green. d Fraction of pathogenic variants that were CNVs in various genes. The top five genes with the most pathogenic variants of any type are shown in each clinical specialty
Genes for hereditary cancer syndromes showed a high (8.3% overall; 0–50% range among panels) prevalence of CNVs among pathogenic variants (Fig. 2a; Supplementary Tables 3 and 4). Among 1059 pathogenic CNVs observed in these genes, 219 were observed only once and 174 were recurrent. BRCA1 and BRCA2 had a combined CNV prevalence of 6.1% (confidence interval [CI]: 5.4–6.9%) among pathogenic variants, consistent with previous studies (individually, BRCA1 11.4% [CI: 10.0–12.9%], BRCA2 1.7% [CI:1.2–2.3%]).15,19,20 CNVs were also enriched in other genes, such as EPCAM, STK11, and VHL, and in genes on various panels with low overall diagnostic yields. Using our NGS method, we also observed 90 CNVs in the segmentally duplicated exons 12–15 of the functional gene copy of PMS2 (Supplementary Table 1). Last, 25 CNVs were observed in promoter regions of GREM1, TP53, and APC.
CNVs in genes associated with pediatric and rare disorders accounted for 7.7% of pathogenic variants (0–82% range among panels; Fig. 2c). We found the highest frequencies of CNVs in panels for early infantile epileptic encephalopathy, Joubert syndrome, tuberous sclerosis, and cerebral cavernous malformations (Supplementary Table 4). The genes most frequently affected by pathogenic CNVs were NF1, NPHP1, and TSC2 (Supplementary Table 3). Among epilepsy genes, we observed CNVs involving UBE3A in 15q13.1 and PRRT2 in 16p11.2, which were likely recurrent cytogenetic rearrangements. We observed lower CNV frequencies in gene panels for ciliopathies, RASopathies, osteogenesis imperfecta, and cystic fibrosis (Supplementary Table 4). Noonan syndrome and chronic pancreatitis panels identified very few or no pathogenic CNVs, although at least 270 individuals were tested and more than 60 pathogenic variants were reported in each panel.
Genes for cardiovascular disorders showed a comparatively lower prevalence of CNVs among pathogenic variants (4.7% overall; 0–16.7% range among panels). The highest frequencies of CNVs occurred in panels for cardiomyopathy and skeletal muscle disease (a subset of the comprehensive cardiomyopathy panel), familial hypercholesterolemia, and Brugada syndrome (Supplementary Table 4). By contrast, very few CNVs were found in panels for arrhythmias (other than Brugada) and aortopathies, whereas the cardiomyopathies panel had the lowest prevalence of pathogenic CNVs. The genes with the highest number of pathogenic CNVs were LDLR, FBN1, PKP2, MYBPC3, and RYR2 (Supplementary Table 3). In some panels yielding apparently high CNV prevalence, most if not all CNVs were in only one or two genes (e.g., ENG and LDLR). Panels for cardiovascular disorders with the highest overall diagnostic yield also had the genes with the highest prevalence of CNVs, except those for arrhythmias and cardiomyopathies, which were depleted of CNVs and in which most positive diagnoses were instead explained by SNVs.
Gene panels for neurological disorders (mostly neuromuscular disorders in our panels) showed the highest prevalence of intragenic CNVs among pathogenic variants (35% overall, 0–100% range among panels; Fig. 2a,c; Supplementary Table 4). This outcome was largely explained by recurrent gene duplication and reciprocal deletion in PMP22, deletions in SMN1, and various CNVs in DMD (Supplementary Table 3; Fig. 2c,d; Supplementary Figure 2). Using a custom NGS method, we found 135 cases of SMN1 deletion among 819 individuals with suspected spinal muscular atrophy, and the range of SMN2 copies varied from 0 to 5. Even when PMP22, SMN1, and DMD were excluded, intragenic CNVs in genes linked to neurological disorders still represented 6% of all pathogenic variants in our cohort. Other genes for neurological disorders commonly affected by CNVs included PARK2, LAMA2, and SPG11.
Analysis of baseline CNVs
Our diagnostic testing was limited to disease genes requisitioned by physicians, but many genes unrelated to the presenting clinical phenotype were also sequenced on our NGS assays. We de-identified data for all 1507 genes sequenced in 143,142 individuals and investigated the occurrence of intragenic CNVs in non-requisitioned genes to estimate the baseline prevalence of these events. These phenotype-independent CNVs are hereafter referred to as “baseline CNVs.” A search for baseline CNVs was performed in 7–616 genes per individual for a total of 16 million single-gene analyses. This search yielded 4054 intragenic CNVs (1465 distinct events) in 3772 individuals across 599 genes (Supplementary Table 5). Most of these CNVs were present only once, but a few were seen 2 to more than 15 times (Fig. 3a; Supplementary Table 6). However, the recurrent events in aggregate accounted for most of the baseline CNV observations. The vast majority of genes with baseline CNVs had five or fewer events (Fig. 3b). A mere 47 genes contained more than half of all observed baseline CNVs, including both genes with identical recurrent events and those with a multitude of unique events. Most individuals with an intragenic baseline CNV had only a single event, but 146 individuals had additional CNVs in genes on different chromosomes. On average, we detected a baseline CNV at a rate of 1 in every 3979 genes sequenced with our assays.
The clinical significance of these CNVs was not evaluated beyond their being deletions in American College of Medical Genetics and Genomics–listed genes with known loss-of-function (LOF) mutational mechanisms. CNVs in genes that belonged to the same clinical specialty as those ordered for clinical testing were removed from the analysis. Single-exon low-quality calls were removed to reduce potential false-positive calls. a Histogram showing the number of distinct CNVs observed in genes analyzed for baseline CNVs. The columns in the chart show the number of times the CNVs were observed. The line graph shows the proportion of total observed CNVs in each frequency bin. For example, the first column shows that more than 1100 CNVs occurred just once and, in aggregate, accounted for roughly 25% of all CNVs. By contrast, the last column indicates that 36 CNVs were found more than 15 times and represented more than 40% of all baseline CNVs. b Histogram showing the number of genes that contained baseline CNVs. The columns in the chart show incremental increases in the number of CNVs observed in a gene. The line graph shows the proportion of CNVs at arbitrary increments of CNV occurrence per gene. For example, nearly 240 genes had just one CNV, and together these CNVs accounted for roughly 6% of all events. By contrast, approximately 45 genes had more than 15 CNVs each, which represented slightly more than 60% of all CNVs. c Distribution of baseline CNVs is shown according to the number of exons affected. Multiexon CNVs include three classes of 5′ terminal exons, internal exons, and 3′ terminal exons. (D and E) Burden of baseline CNVs in genes according to mode of inheritance and zygosity. CNVs in genes with LOF mutational mechanisms are shown in part (d) and those in genes without LOF mechanisms are shown in part (e). CNVs in X-linked (XL) genes were categorized as those observed as heterozygous events in females and hemizygous events in males. AD autosomal dominant, AR autosomal recessive, F female, M male, het heterozygous, hom homozygous
In contrast to CNVs identified in the clinically tested genes in this cohort, most baseline intragenic CNVs were duplications (Figs. 1c,d and 3c). Most were also heterozygous variants in AR genes or genes that lacked established LOF mechanisms (Fig. 3d,e). A minority of baseline CNVs occurred in genes associated with AD inheritance or LOF mechanisms (Figs. 1e,f and 3d,e). The most common baseline CNVs included whole-gene events in NPHP1, NIPA1, MYH11, DNAI2, HFE2, SMN1, and PMP22 and partial-gene events in TFG, BBS9, CTNNA3, PARK2, KCTD7, DNAJC6, GLIS2, and TUBB4A (Supplementary Table 6). In terms of characteristics that may explain the existence of baseline CNVs in disease genes, we noted that nearly 40% of these CNVs encompassed a whole gene and therefore did not directly disrupt transcript reading frames (Fig. 3c). Furthermore, roughly 90% of the duplications in genes with LOF mechanisms were whole-gene events or partial-gene events including a terminal exon, whereas only half of the deletions in these genes showed the same patterns (Supplementary Table 5).
In addition to assessing the overall prevalence and properties of baseline CNVs, we considered predicted clinical implications. We observed 237 heterozygous deletions in 97 genes with AD or XL inheritance and LOF mechanisms; most were in PMP22, DMD, AARS, KCNQ1, FIG4, CHEK2, and LRSAM1 (Supplementary Tables 5 and 7). We found only two homozygous deletions in genes with AR inheritance (NPHP1 and SPG7) and only two hemizygous deletions in a single gene with XL inheritance (DMD) in males. All other homozygous CNVs in genes with AR inheritance, or hemizygous CNVs in genes with XL inheritance in males, were duplications. Furthermore, we observed CNVs specifically in genes with medical actionability considerations according to the ACMG.21,22 We evaluated CNVs in 58 of the 59 ACMG-listed genes (excluding PMS2) in 5,300–69,000 individuals depending on the assays used for testing. A total of 46 deletions and 110 duplications were detected, suggesting a frequency of up to 0.8% (CI: 0.58–1.11%) among individuals tested for those genes. MYH11, MYH7, KCNQ1, and RYR2 contained most of the CNVs. Specifically, there were deletions in 16 genes—KCNQ1, MYH11, MYH7, MYBPC3, PCSK9, BRCA1, RYR2, PKP2, TGFBR2, SMAD3, OTC, NF2, FBN1, DSP, DSC2, and APC—more than half of which have LOF mechanisms (Supplementary Table 7).
Discussion
Prevalence of clinically important intragenic CNVs
The NGS gene panel data we describe show that although individual intragenic CNVs may be rare, they constitute a substantial proportion of clinically important variants in aggregate. Approximately 60% of the CNVs in our cohort were individually rare; half of these were found just once, and the remainder occurred no more than five times. CNVs accounted for ~3% of all reported variants but a notable ~9% of clinically significant LP/P variants in different gene panels across clinical specialties. This observation emphasizes the importance of using high-resolution, exon-level copy number detection methods broadly in genetic testing.
CNVs in relation to gene structure
The location of a CNV within an individual gene can determine how it alters the structure of that gene and therefore influences its clinical interpretation. Among CNVs involving the first or last terminal exons, more deletions than duplications included first exons. This outcome was expected in our cohort because most of the genes we tested had LOF mechanisms and deletions at the 5′ ends of genes are almost invariably damaging. By contrast, the number of partial-gene deletions and duplications including 3′ terminal exons were comparable, which suggests that these CNVs may contribute less to morbidity. Similarly, partial-gene duplications involving 3′ terminal exons and full-gene duplications may not always be deleterious because they can leave functional gene structure intact (Fig. 1d). Indeed, these types of CNVs are present in disease genes (e.g., CREBBP) in DGV, which has substantial data on CNVs in the general population.14 Therefore, partial-gene CNVs involving 3′ terminal exons may constitute a proportion of normal variation in the genome and should be scrutinized carefully when detected during genetic testing. On the contrary, partial-gene CNVs that involve only internal exons likely disrupt the transcript reading frame and explain disease. As expected, these constituted the majority in our clinical cohort. However, we classified some CNVs involving only internal exons as VUS because their effect on the transcriptional reading frame was unclear. Indeed, some of these were also listed as low-frequency events (e.g., in LRSAM1, NRXN1, and SCN9A) in healthy individuals in ExAc and DGV.3,6
Patterns of CNV occurrence
Some genes have a single recurrent CNV, others have a multitude of distinct CNVs, and still others include a mix of both. Common CNVs affecting an entire gene tend to occur in genes such as PMP22, MYH11, NPHP1, PRRT2, and UBE3A, which exist within chromosomal regions that undergo recurrent rearrangements.23 Some partial-gene CNVs are also recurrent but less common, as in BRCA1, RYR2, and VHL. However, most disease genes appear to have nonrecurrent, rare, and random intragenic CNVs. For instance, we identified CNVs that in aggregate affected every exon in DMD and involved a broad variety of exon combinations (Supplementary Figure 2). A small number of genes, often involving AD disorders with reduced penetrance, had both nonrecurrent and recurrent CNVs. For example, we observed 74 distinct CNVs in BRCA1, of which 36 occurred only once and 11 occurred five or more times and included founder mutations segregating on specific haplotypes within ethnic groups.24,25
Intragenic CNVs across clinical specialties
Panels for cancer syndromes, pediatric and rare disorders, and neurological disorders showed a high CNV prevalence overall. For instance, an expected CNV prevalence of 6–22% among pathogenic variants was observed in BRCA1 and BRCA2 testing for hereditary breast and ovarian cancer or VHL testing for von Hippel–Lindau disease.15,20,26 The CNV prevalence in panels for pediatric and neurological disorders would be higher had we included testing for classic developmental syndromes and intellectual disability disorders that are often caused by CNVs detectable on chromosomal microarrays.27 By contrast, panels for cardiovascular disorders showed a lower CNV prevalence, likely because these disorders are predominantly caused by gain-of-function mechanisms that typically act through SNVs rather than CNVs. Consistent with our results, a recent study of cardiomyopathies also showed a low rate of CNVs.28
Baseline CNVs as naturally existing variation
CNVs in clinically relevant genes are mostly deletions and partial-gene events and often involve only the internal exons of genes. These properties distinguish them from baseline CNVs, which mostly exist as duplications and full-gene events and predominantly involve terminal 5′ and 3′ exons. Furthermore, baseline CNVs are largely sequestered in genes associated with disorders that exhibit AR inheritance, AD inheritance with reduced penetrance, or AD inheritance and gain-of-function mechanisms.
Baseline CNVs can be divided into two categories—those that have a likely benign effect because they do not adversely affect the structural integrity of a gene and those that are pathogenic but present only as latent carrier alleles. Less than 10% of baseline CNVs in our cohort were in genes associated with dominant, highly penetrant, early-onset disorders and had properties of CNVs in the first category. We also identified a small number of baseline CNVs in the second category. For example, we found 23 cases of PMP22 duplication, which mirrors findings in a recent report of intragenic CNVs. Similarly, we identified duplications involving TBX1 in 22q11.21, HFE2 in 1q21.1, and PRRT2 in 16p11.2; these were not unexpected because they are part of well-characterized cytogenetic microduplications with reduced penetrance.29 In addition, most of the CNVs in genes prescribed by the ACMG for reporting secondary findings were duplications. However, there were deletions in at least 16 genes, of which more than half have LOF mechanisms. Our data indicate that up to 0.8% of individuals in the general population may have additional CNVs in the ACMG-listed genes. Because studies of secondary findings have focused almost exclusively on SNVs or other sequence changes,30,31 data on intragenic CNVs in the ACMG-listed genes provide useful complementary insight into disease-predisposing variants observable in healthy individuals.32
Study limitations
Because we evaluated only 1507 disease genes and excluded non–disease genes, the catalog of intragenic CNVs in the human genome requires further investigation. We were also unable to test every gene in our clinical menu in every individual because only assays containing the clinician-ordered diagnostic panel were used for each individual. Therefore, some genes were tested much more frequently than others, and CNV data from these genes should be interpreted in that context. Furthermore, the majority of the genes we tested are involved in disorders of haploinsufficiency and therefore are expected to have more CNVs with LOF effects than would be the case if a random set of genes were analyzed. Finally, our assays lacked the resolution required to identify precise breakpoints for most CNVs. Genome sequencing will soon provide the clarity needed to address some of these limitations and to understand the mechanisms that create intragenic CNVs.
This study is among the first to investigate the prevalence and properties of intragenic CNVs across >1000 disease genes in a large clinical cohort. The ~10% prevalence of CNVs among individuals with a positive test result is a novel observation that emphasizes the need to routinely evaluate intragenic copy number in genetic testing. The data presented here also bridge a long-standing gap in molecular genetic analysis between SNVs and indels at one end of the spectrum of detectable variants and chromosomal CNVs at the other, thereby providing a more complete picture of the molecular etiologies responsible for hereditary disease. Finally, in addition to cultivating a better appreciation of the involvement of CNVs in clinical phenotypes, these data provide insight into baseline CNVs, thereby enhancing the resolution of the human genome dosage map and further illuminating CNV variation that exists naturally in the general population.
References
Pfundt R, Del Rosario M, Vissers L, et al. Detection of clinically relevant copy-number variants by exome sequencing in a large cohort of genetic disorders. Genet Med. 2017;19:667–75.
Retterer K, Scuffins J, Schmidt D, et al. Assessing copy number from exome sequencing and exome array CGH based on CNV spectrum in a large clinical cohort. Genet Med. 2015;17:623–9.
Aradhya S, Lewis R, Bonaga T, et al. Exon-level array CGH in a large clinical cohort demonstrates increased sensitivity of diagnostic testing for Mendelian disorders. Genet Med. 2012;14:594–603.
Gambin T, Yuan B, Bi W, et al. Identification of novel candidate disease genes from de novo exonic copy number variants. Genome Med. 2017;9:83.
Dhami P, Coffey AJ, Abbs S, et al. Exon array CGH: detection of copy-number changes at the resolution of individual exons in the human genome. Am J Hum Genet. 2005;76:750–62.
Hehir-Kwa JY, Pfundt R, Veltman JA. Exome sequencing and whole genome sequencing for the detection of copy number variation. Expert Rev Mol Diagn. 2015;15:1023–32.
Johansson LF, van Dijk F, de Boer EN, et al. CoNVaDING: single exon variation detection in targeted NGS data. Hum Mutat. 2016;37:457–64.
Chen Y, Zhao L, Wang Y, et al. SeqCNV: a novel method for identification of copy number variations in targeted next-generation sequencing data. BMC Bioinformatics. 2017;18:147.
Lincoln SE, Kobayashi Y, Anderson MJ, et al. A systematic comparison of traditional and multigene panel testing for hereditary breast and ovarian cancer genes in more than 1000 patients. J Mol Diagn. 2015;17:533–44.
Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Sudmant PH, Rausch T, Gardner EJ, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81.
Mills RE, Walter K, Stewart C, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65.
Landrum MJ, Lee JM, Benson M, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–8.
Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16:172–83.
Kurian AW, Hare EE, Mills MA, et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol. 2014;32:2001–9.
Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Nykamp K, Anderson M, Powers M, et al. Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet Med. 2017;19:1105–17.
Newman S, Hermetz KE, Weckselblatt B, Rudd MK. Next-generation sequencing of duplication CNVs reveals that most are tandem and some create fusion genes at breakpoints. Am J Hum Genet. 2015;96:208–20.
Judkins T, Rosenthal E, Arnell C, et al. Clinical significance of large rearrangements in BRCA1 and BRCA2. Cancer. 2012;118:5210–6.
Petrucelli N, Daly MB, Pal T. BRCA1- and BRCA2-associated hereditary breast and ovarian cancer. In: MP Adam, HH Ardinger, RA Pagon, et al., editors. GeneReviews. Seattle, WA: University of Washington; 1993.
Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–74.
Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SFv2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–55.
Vissers LE, Stankiewicz P. Microdeletion and microduplication syndromes. Methods Mol Biol. 2012;838:29–75.
The BRCA1 Exon 13 Duplication Screening Group. The exon 13 duplication in the BRCA1 gene is a founder mutation present in geographically diverse populations. Am J Hum Genet. 2000;67:207–12.
Weitzel JN, Lagos VI, Herzog JS, et al. Evidence for common ancestral origin of a recurring BRCA1 genomic rearrangement identified in high-risk Hispanic families. Cancer Epidemiol Biomark Prev. 2007;16:1615–20.
Frantzen C, Klasson, TD, Links, TP, and Giles, RH (1993). Von Hippel-Lindau syndrome. In: MP Adam, HH Ardinger, RA Pagon, et al., editors. GeneReviews. Seattle, WA: University of Washington; 1993.
Martin CL, Kirkpatrick BE, Ledbetter DH. Copy number variants, aneuploidies, and human disease. Clin Perinatol. 2015;42:227–42. vii
Ceyhan-Birsoy O, Pugh TJ, Bowser MJ, et al. Next generation sequencing-based copy number analysis reveals low prevalence of deletions and duplications in 46 genes associated with genetic cardiomyopathies. Mol Genet Genom Med. 2016;4:143–51.
Rosenfeld JA, Coe BP, Eichler EE, Cuckle H, Shaffer LG. Estimates of penetrance for recurrent pathogenic copy-number variations. Genet Med. 2013;15:478–81.
Olfson E, Cottrell CE, Davidson NO, et al. Identification of medically actionable secondary findings in the 1000 Genomes. PLoS ONE. 2015;10:e0135193.
O’Daniel JM, McLaughlin HM, Amendola LM, et al. A survey of current practices for genomic sequencing test interpretation and reporting processes in US laboratories. Genet Med. 2017;19:575–82.
Innes J, Reali L, Clayton-Smith J, et al. CNVs affecting cancer predisposing genes (CPGs) detected as incidental findings in routine germline diagnostic chromosomal microarray (CMA) testing. J Med Genet. 2017;55:89–96.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
All authors are employees of Invitae, a laboratory offering clinical genetic testing services.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Truty, R., Paul, J., Kennemer, M. et al. Prevalence and properties of intragenic copy-number variation in Mendelian disease genes. Genet Med 21, 114–123 (2019). https://doi.org/10.1038/s41436-018-0033-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-018-0033-5
Keywords
This article is cited by
-
Detection of a novel gross deletion in the UNC13D gene ends the diagnostic odyssey for a family with familial hemophagocytic lymphohistiocytosis 3
BMC Pediatrics (2024)
-
BARD1 deletion in a patient with suspected hereditary colorectal cancer
Human Genome Variation (2024)
-
Intragenic homozygous duplication in HEPACAM is associated with megalencephalic leukoencephalopathy with subcortical cysts type 2A
neurogenetics (2024)
-
Whole-genome sequencing of 1029 Indian individuals reveals unique and rare structural variants
Journal of Human Genetics (2023)
-
Identification of variants in genes associated with hypertrophic cardiomyopathy in Mexican patients
Molecular Genetics and Genomics (2023)