Abstract
For nearly a decade, our research group has had the privilege of developing and mining a multicenter, microarray-based, genome-wide expression database of critically ill children (≤10 y of age) with septic shock. Using bioinformatic and systems biology approaches, the expression data generated through this discovery-oriented, exploratory approach have been leveraged for a variety of objectives, which are reviewed here. Fundamental observations include widespread repression of gene programs corresponding to the adaptive immune system and biologically significant differential patterns of gene expression across developmental age groups. The data have also identified gene expression–based subclasses of pediatric septic shock having clinically relevant phenotypic differences. The data have also been leveraged for the discovery of novel therapeutic targets, as well as for the discovery and development of novel stratification and diagnostic biomarkers. Almost a decade of genome-wide expression profiling in pediatric septic shock is now demonstrating tangible results. The studies have progressed from an initial discovery-oriented and exploratory phase to a new phase in which the data are being translated and applied to address several areas of clinical need.
Similar content being viewed by others
Main
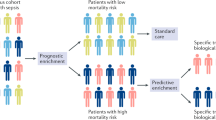
For nearly a decade, our research group has had the privilege of developing and mining a multicenter, microarray-based, genome-wide expression database of critically ill children (≤10 y of age) with septic shock. The expression data are based on whole blood–derived RNA and are focused on the initial, acute presentation to the pediatric intensive care unit (PICU) with a clinical diagnosis of septic shock. Comparator groups include age-matched normal controls, critically ill children meeting criteria for sepsis, and critically ill children meeting criteria for the systemic inflammatory response syndrome (SIRS), based on pediatric-specific definitions (1). Using bioinformatic and systems biology approaches, the expression data generated through this discovery-oriented, exploratory approach have been leveraged for a variety of objectives, which are summarized in Figure 1 and are reviewed below. Analogous studies involving adult populations were recently reviewed (2).
Genome-Level Response of Pediatric Septic Shock and Repression of Adaptive Immunity–Related Genes
The long-standing paradigm for understanding the pathophysiology of septic shock is centered on a dysfunctional innate immune system, wherein excessive and uncontrolled proinflammatory responses lead to direct tissue and organ injury (3,4). Although this paradigm remains valid, the vast majority of clinical trials focused on controlling or modulating this excessive inflammatory response have failed to demonstrate efficacy, thereby leading to the derivation of alternative paradigms for understanding the pathophysiology of human sepsis (5). One such alternative paradigm focuses on dysfunction of the adaptive immune system, wherein a form of immune suppression or “immune paralysis” accounts for the negative consequences of sepsis (6,7,8,9,10).
Genome-level expression patterns in children with septic shock strongly support this concept of immune suppression (2,11,12,13,14,15,16,17). Specifically, pediatric septic shock is characterized by widespread repression of gene programs corresponding to various major components of the adaptive immune system, including the T-cell receptor signaling pathway, T-cell function, B-cell function, and the major histocompatibility complex antigen presentation pathway. This pattern of gene repression is evident within the first 24 h of presentation to the pediatric intensive care unit (PICU) with septic shock and persists at least into the third day of PICU admission. Analogous studies in adults with septic shock, and in adults suffering from major traumatic injuries, have also demonstrated similar patterns of gene repression corresponding to the adaptive immune system (18,19,20,21,22). Accordingly, novel therapeutic strategies for septic shock are now being contemplated with a focus on restoration of the adaptive immune system, rather than inhibition of the innate immune system and the inflammatory response (23,24).
Influence of Development on the Genome-Level Response of Pediatric Septic Shock
A variety of clinical and experimental data indicate that developmental age influences the immune system and hence the host response to sepsis (4,25,26,27,28). A recent study examined the acute, genome-level expression patterns across four developmental age groups with septic shock: neonate (≤28 d of age), infant (1 mo through 1 y of age), toddler (2–5 y of age), and school age (≥6 y of age) (17).
In comparisons with age-matched controls, the neonate and school-age groups had the largest number of uniquely regulated genes corresponding to several key inflammation- and immunity-related pathways. In direct comparisons across the developmental age groups, there were minimal differences among the infant, toddler, and school-age groups. In contrast, age-specific differential gene expression was most profound in the neonate group, which predominately demonstrated reduced expression of several key pathways of both the innate and the adaptive immune response. This observation does not appear to be an artifact of differential white blood cell counts. The expression data were analyzed for the presence of signature probe sets for neutrophils, lymphocytes, and monocytes. The signature probe sets were present to a similar degree across the four developmental age groups, thereby suggesting that the relative contributions of the three major leukocyte subsets to the gene expression profiles did not substantially differ across the four developmental age groups.
One striking example involved expression of genes related to the triggering receptor expressed on myeloid cells-1 signaling pathway, for which there has been recent interest as a therapeutic target in sepsis (29). Genes corresponding to the triggering receptor expressed on myeloid cells-1 signaling pathway were not expressed in the neonate group, whereas they were highly expressed in the three older developmental age groups. These data illustrate how development must be taken into consideration when developing novel, immunity-based therapies for sepsis.
Discovery of Gene Expression–Based Subclasses
Septic shock is a heterogeneous syndrome, rather than a discrete, uniform disease process (5). This high level of heterogeneity implies the potential existence of disease “subclasses” predicated on distinct patterns of gene expression. A series of three recent studies indicate that gene expression–based subclasses of pediatric septic shock may indeed exist and that the subclasses have clinically important phenotypic differences (30,31,32).
The first study in this series attempted to identify subclasses of pediatric septic shock based exclusively on differential patterns of gene expression (30). A list of more than 6,000 differentially regulated genes was generated using statistical and expression filters targeted at subclass discovery. The differentially regulated genes were then subjected to unsupervised hierarchical clustering, and three putative gene expression–based subclasses (subclasses A, B, and C) were identified based on the first- and second-order cluster branches. Post hoc analysis of the gene expression–based subclasses revealed that patients in subclass A had a significantly higher degree of illness severity and a significantly higher mortality rate compared with patients in subclasses B and C.
The subclass-defining gene signature was subsequently distilled to the top 100 class predictor genes, and these genes corresponded to T-cell receptor signaling, B-cell receptor signaling, glucocorticoid-receptor signaling, and peroxisome proliferator–activated receptor-α activation (30). Of note, the genes corresponding to these signaling pathways were generally repressed in the subclass A patients, who also had the highest degree of illness severity and the highest mortality rate among the three identified subclasses. Recently, these observations were experimentally corroborated by the demonstration that peroxisome proliferator–activated receptor-α–deficient mice are more susceptible to the untoward effects of experimental sepsis (33).
In the second study of this series, the 100 class-defining genes were depicted using gene expression mosaics that allow for intuitive pattern recognition of otherwise complex gene expression patterns (32,34,35). Clinicians with no formal bioinformatic training and minimal instruction were able to allocate patients to the correct gene expression–based subclasses with a high degree of accuracy, thus demonstrating the potential feasibility of bringing gene expression–based subclassification of pediatric septic shock to the bedside. In the final study of this series, the gene expression mosaics were used to allocate a new, test cohort of patients into each of the three putative subclasses (31). This validation study corroborated that patients in subclass A have a higher degree of illness severity compared with patients in subclasses B and C. Collectively, this series of studies indicates that gene expression–based subclasses of pediatric septic shock exist, that the subclasses can be readily identified by clinicians using gene expression mosaics, that the class-defining genes are biologically relevant, and that the subclasses have clinically relevant phenotypic differences.
Discovery of Novel Therapeutic Targets
A common goal for discovery-oriented clinical studies centered on genome-wide expression patterns involves the discovery of novel therapeutic targets. The pediatric genome-wide expression studies have thus far identified two promising areas for therapeutic intervention, which have been corroborated experimentally: inhibition of matrix metallopeptidase-8 (MMP-8) activity and restoration of zinc homeostasis.
MMP-8 belongs to a family of 25 individual endopeptidases having different cellular sources, expression conditions, and target substrates (36). MMP-8 is also known as neutrophil collagenase given its original identification as a neutrophil product that remodels the extracellular matrix via cleavage of collagen type I. However, it is now apparent that multiple cell types have the ability to express MMP-8, particularly in the setting of inflammatory conditions. In addition, MMP-8 can also cleave a wide range of noncollagenous substrates, including serine protease inhibitors and several chemokines.
MMP-8 is consistently the highest expressed gene in children with septic shock, and the degree of expression and proteolytic activity positively correlate with the degree of illness severity and mortality, but do not correlate with differential white blood cell counts (37). Similar observations have been reported in adults with sepsis (38). These observations have led to a series of experiments designed to formally test the role of MMP-8 in a murine model of sepsis (37). MMP-8 null mice have a significant survival advantage in experimental sepsis compared with wild-type animals. This survival advantage correlates with an attenuated inflammatory response, without compromising clearance of bacteria. In addition, this phenotype can be replicated by treating wild-type mice with a pharmacologic inhibitor of MMP-8 activity, after induction of sepsis. Collectively, these data indicate that MMP-8 inhibition is a potential, novel therapeutic strategy for sepsis. This assertion is supported by the availability of clinically approved pharmaceutical compounds that effectively inhibit MMP-8 activity (36,39).
Zinc is an essential trace element and is critical for normal functioning of both the innate and the adaptive immune systems (40). A consistent observation across multiple gene expression studies is that pediatric septic shock is characterized by widespread repression of gene families that either directly participate in zinc homeostasis or are directly dependent on zinc homeostasis for normal function (11,12,14,15,16,30). These observations suggest that altered zinc homeostasis may be a prominent feature of pediatric septic shock. Indeed, low serum zinc concentrations have been documented in pediatric septic shock, and nonsurvivors have lower serum zinc concentrations compared with survivors (16). Low serum zinc concentrations have also been reported in other forms of critical illness in both adults and children (41,42,43).
These observations have collectively fostered the concept that zinc supplementation may be a low-cost and effective therapeutic strategy for septic shock (41,44). In animal models of sepsis, zinc supplementation confers a survival benefit, and the survival benefit correlates with enhanced bacterial clearance as well as modulation of excessive inflammation (45,46,47). Two recent studies involving children with either pneumonia or bacteremia in developing countries demonstrated that zinc supplementation significantly reduced infection-related complications, including mortality (48,49). A study involving critically ill children in the United States tested the efficacy of oral zinc supplementation, in combination with oral selenium, glutamine, and metoclopramide, as a means of preventing nosocomial infection or sepsis (24). There was no efficacy in the overall study population for the primary study end point (time to development of nosocomial infection/sepsis), but a secondary analysis restricted to patients with baseline immune dysfunction suggested a beneficial effect in reducing nosocomial infections. Thus, zinc supplementation as an adjunctive therapeutic strategy for septic shock remains an intriguing concept that requires further testing. There are currently two ongoing phase 1 trials of intravenous zinc supplementation designed to address this question further (ClinicalTrials.gov: NCT01062009 and NCT01162109).
Discovery of Stratification Biomarkers
The ability to stratify outcome risk is fundamental to effective clinical practice and clinical research, and there is great deal of interest in using biomarkers to stratify outcome risk in septic shock (5,50,51,52). Effective stratification biomarkers could benefit the field of septic shock in three broad areas: risk stratification for clinical trials, informing decision making for individual patients, and as a metric for quality improvement efforts. The genome-wide expression studies in pediatric septic shock have served as effective tools for the discovery of candidate stratification biomarkers.
The first genome-wide expression study in pediatric septic shock generated a list of genes differentially expressed between survivors and nonsurvivors, thus providing an initial working list of candidate stratification biomarkers (16). Interleukin-8 (IL-8), the principal human chemoattractant for neutrophils, was among these differentially expressed genes; validation assays confirmed that serum IL-8 protein levels were indeed increased in nonsurvivors compared with survivors (16). This observation led to a subsequent study directly testing the ability of serum IL-8 levels to predict 28-d mortality vs. survival (53). This study demonstrated that an IL-8 level ≤220 pg/ml, within 24 h of presentation to the PICU with septic shock, had a negative predictive value for mortality of 95%. This observation was subsequently validated in an independent test cohort. Consequently, it has been proposed that serum IL-8 levels can be used to exclude low-risk patients from future pediatric septic shock interventional trials as a means to improve the risk-to-benefit ratio of a therapeutic intervention. Of note, IL-8 does not seem to be an effective stratification biomarker in adult septic shock, thereby suggesting important biological and physiological differences between adult and pediatric septic shock (54).
Although the negative predictive value of serum IL-8 alone is high, the other test characteristics of IL-8 (i.e., sensitivity, specificity, and positive predictive value) are not sufficiently robust to develop a comprehensive pediatric septic shock stratification tool meeting a wide variety of clinical and research needs. Given the complexity of septic shock, it is possible that a multibiomarker-based stratification system could better address these needs. Accordingly, the genome-wide expression data in pediatric septic shock have been leveraged to select, objectively, a panel of 12 candidate stratification biomarkers (51). Using classification and regression tree methodology, the candidate stratification biomarkers have been used to derive and test a decision tree–based model that robustly predicts outcome in children with septic shock (55). The model has been tested in 355 subjects and has the following test characteristics for predicting 28-d all-cause mortality: sensitivity 93%, specificity 74%, positive predictive value 32% (in the context of an overall mortality rate of ~10%), and negative predictive value 99%. Although the model requires further testing, the test characteristics indicate that it has substantial potential to serve a wide variety of stratification needs in pediatric septic shock.
Discovery of Diagnostic Biomarkers
The development of diagnostic biomarkers is another area of interest in the field of pediatric critical care medicine, and again, genome-wide expression data have been leveraged for the discovery of two classes of candidate diagnostic biomarkers: biomarkers to diagnose the development of septic shock–associated kidney injury and biomarkers to diagnose the presence of bacterial infection in critically ill children.
Acute kidney injury (AKI) is a common complication of critical illness and septic shock; and consequently, there is substantial interest in the development of biomarkers that diagnose AKI before a rise in serum creatinine as a means to institute potentially renal protective therapies in a more timely manner (56). Commonly used biomarkers for AKI may not be as effective in the context of septic shock compared with other etiologies of AKI (56,57,58). Accordingly, there remains a need to develop AKI diagnostic biomarkers specific to septic shock.
Genome-wide expression data have been leveraged for the discovery of 21 unique genes that can predict the development of septic shock–associated kidney injury with a sensitivity of 98% and a specificity of 80% (59). The products of several of these genes (i.e., proteins) can be readily measured in the serum compartment, and preliminary data indicate that the serum protein concentrations can also predict the development of septic shock–associated kidney injury (59). Studies are currently in progress to determine whether a panel of these candidate, serum-based, diagnostic markers can improve the ability to diagnose septic shock–associated kidney injury further.
A daily clinical conundrum in the PICU is the differentiation of SIRS secondary to infection (i.e., sepsis) from SIRS secondary to noninfectious etiologies (i.e., “sterile inflammation”). Procalcitonin has emerged as a potential diagnostic biomarker to meet this need, but its performance in critically ill populations has been challenged (60).
The most recent analysis of the pediatric genome-wide expression data has been targeted toward the identification of diagnostic biomarkers to identify the presence of bacterial infection in critically ill children (61). The analysis compared critically ill children with SIRS having documented negative bacterial cultures (i.e., the noninfected group) with those having documented positive bacterial cultures (i.e., the infected group). A total of 221 gene probes were differentially regulated between the noninfected group and the infected group. A leave-one-out crossvalidation procedure based on a Support Vector Machines learning algorithm, and the 221 differentially regulated gene probes were able to differentiate the two groups with 86% accuracy. Epstein–Barr virus–induced gene 3 had the highest predictive strength among these 221 gene probes.
Epstein–Barr virus–induced gene 3 is a subunit of the heterodimeric cytokine IL-27, and IL-27 has been recently linked to the pathophysiology of sepsis (62,63). Because IL-27 can be readily measured in the serum compartment, the ability of IL-27 to diagnose bacterial infection in critically ill children was tested and compared with that of procalcitonin (61). Serum IL-27 levels had a >90% specificity and positive predictive value for diagnosing bacterial infection in this cohort of critically ill children, and the overall performance of IL-27 was generally better than that of procalcitonin. In addition, a decision tree incorporating IL-27 and procalcitonin performed better than either biomarker alone. Thus, IL-27 has potential as an effective “rule-in” test for bacterial infection in critically ill children. Additional studies are being planned to test further the ability of IL-27 to serve as a biomarker for bacterial infection.
Potential Limitations
The majority of the studies reviewed above are based on whole blood–derived RNA. Thus, the source of RNA is a mixed population of white blood cells. Although this approach has facilitated the accumulation of a large number of biological samples from multiple institutions, it has the potential to confound data inasmuch as differential patterns of gene expression could reflect differential white blood cell counts, rather than intrinsic differences in mRNA expression. This potential confounder has been addressed by multiple complementary approaches, including corrections for differential white blood cell counts (11,12), interrogation of gene expression data for leukocyte subset–specific gene probes (15,17,64), the generation of expression data using leukocyte subset–specific mRNA (15), and the ability to validate gene expression signatures in formal validation cohorts (11,31). In addition, the gene expression data generated through these studies have been successfully leveraged for the development of serum protein biomarkers (51–53,55,59,65) and have been validated in animal models of sepsis (33,37,47). Thus, although the potential confounder of using whole blood–derived RNA remains an important issue, it would appear that the data generated thus far primarily reflect intrinsic differences in gene expression rather than an artifact of mRNA derived from a mixed population of white blood cells.
Conclusion
Almost a decade of genome-wide expression profiling in pediatric septic shock is now demonstrating tangible results. The studies have progressed from an initial discovery-oriented and exploratory phase to a new phase in which the data are being translated and applied to address several areas of clinical need. These areas include the identification of potential novel therapeutic targets and pathways, discovery of candidate diagnostic and stratification biomarkers, and the possibility of clinically relevant and clinically feasible gene expression–based subclassification. The challenges moving ahead include robust validation studies, which will require large-scale collaborations.
Statement of Financial Support
This study was supported by National Institutes of Health grants R01GM099773, R01GM096994, and RC1HL100474.
Disclosures
The Cincinnati Children’s Hospital Research Foundation and the author have submitted provisional patent applications for themes discussed in the manuscript. They include interleukin-8 as a stratification biomarker, a multibiomarker–based stratification model, and interleukin-27 as a diagnostic biomarker.
References
Goldstein B, Giroir B, Randolph A; International Consensus Conference on Pediatric Sepsis. International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med 2005;6:2–8.
Wong HR . Clinical review: sepsis and septic shock–the potential of gene arrays. Crit Care 2012;16:204.
Cornell TT, Wynn J, Shanley TP, Wheeler DS, Wong HR . Mechanisms and regulation of the gene-expression response to sepsis. Pediatrics 2010;125:1248–58.
Wynn J, Cornell TT, Wong HR, Shanley TP, Wheeler DS . The host response to sepsis and developmental impact. Pediatrics 2010;125:1031–41.
Marshall JC . Sepsis: rethinking the approach to clinical research. J Leukoc Biol 2008;83:471–82.
Boomer JS, To K, Chang KC, et al. Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA 2011;306:2594–605.
Hotchkiss RS, Opal S . Immunotherapy for sepsis–a new approach against an ancient foe. N Engl J Med 2010;363:87–9.
Felmet KA, Hall MW, Clark RS, Jaffe R, Carcillo JA . Prolonged lymphopenia, lymphoid depletion, and hypoprolactinemia in children with nosocomial sepsis and multiple organ failure. J Immunol 2005;174:3765–72.
Hall MW, Knatz NL, Vetterly C, et al. Immunoparalysis and nosocomial infection in children with multiple organ dysfunction syndrome. Intensive Care Med 2011;37:525–32.
Hotchkiss RS, Coopersmith CM, McDunn JE, Ferguson TA . The sepsis seesaw: tilting toward immunosuppression. Nat Med 2009;15:496–7.
Cvijanovich N, Shanley TP, Lin R, et al.; Genomics of Pediatric SIRS/Septic Shock Investigators. Validating the genomic signature of pediatric septic shock. Physiol Genomics 2008;34:127–34.
Shanley TP, Cvijanovich N, Lin R, et al. Genome-level longitudinal expression of signaling pathways and gene networks in pediatric septic shock. Mol Med 2007;13:495–508.
Wong HR . Genetics and genomics in pediatric septic shock. Crit Care Med 2012;40:1618–26.
Wong HR, Cvijanovich N, Allen GL, et al.; Genomics of Pediatric SIRS/Septic Shock Investigators. Genomic expression profiling across the pediatric systemic inflammatory response syndrome, sepsis, and septic shock spectrum. Crit Care Med 2009;37:1558–66.
Wong HR, Freishtat RJ, Monaco M, Odoms K, Shanley TP . Leukocyte subset-derived genomewide expression profiles in pediatric septic shock. Pediatr Crit Care Med 2010;11:349–55.
Wong HR, Shanley TP, Sakthivel B, et al.; Genomics of Pediatric SIRS/Septic Shock Investigators. Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics 2007;30:146–55.
Wynn JL, Cvijanovich NZ, Allen GL, et al. The influence of developmental age on the early transcriptomic response of children with septic shock. Mol Med 2011;17:1146–56.
Tang BM, Huang SJ, McLean AS . Genome-wide transcription profiling of human sepsis: a systematic review. Crit Care 2010;14:R237.
Tang BM, McLean AS, Dawes IW, Huang SJ, Cowley MJ, Lin RC . Gene-expression profiling of gram-positive and gram-negative sepsis in critically ill patients. Crit Care Med 2008;36:1125–8.
Tang BM, McLean AS, Dawes IW, Huang SJ, Lin RC . The use of gene-expression profiling to identify candidate genes in human sepsis. Am J Respir Crit Care Med 2007;176:676–84.
Tang BM, McLean AS, Dawes IW, Huang SJ, Lin RC . Gene-expression profiling of peripheral blood mononuclear cells in sepsis. Crit Care Med 2009;37:882–8.
Xiao W, Mindrinos MN, Seok J, et al.; Inflammation and Host Response to Injury Large-Scale Collaborative Research Program. A genomic storm in critically injured humans. J Exp Med 2011;208:2581–90.
Unsinger J, McGlynn M, Kasten KR, et al. IL-7 promotes T cell viability, trafficking, and functionality and improves survival in sepsis. J Immunol 2010;184:3768–79.
Carcillo JA, Dean JM, Holubkov R, et al.; Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Collaborative Pediatric Critical Care Research Network (CPCCRN). The randomized comparative pediatric critical illness stress-induced immune suppression (CRISIS) prevention trial. Pediatr Crit Care Med 2012;13:165–73.
Wynn JL, Levy O . Role of innate host defenses in susceptibility to early-onset neonatal sepsis. Clin Perinatol 2010;37:307–37.
Wynn JL, Scumpia PO, Winfield RD, et al. Defective innate immunity predisposes murine neonates to poor sepsis outcome but is reversed by TLR agonists. Blood 2008;112:1750–8.
Cuenca AG, Wynn JL, Kelly-Scumpia KM, et al. Critical role for CXC ligand 10/CXC receptor 3 signaling in the murine neonatal response to sepsis. Infect Immun 2011;79:2746–54.
Wynn JL, Scumpia PO, Delano MJ, et al. Increased mortality and altered immunity in neonatal sepsis produced by generalized peritonitis. Shock 2007;28:675–83.
Bouchon A, Facchetti F, Weigand MA, Colonna M . TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature 2001;410:1103–7.
Wong HR, Cvijanovich N, Lin R, et al. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med 2009;7:34.
Wong HR, Cvijanovich NZ, Allen GL, et al. Validation of a gene expression-based subclassification strategy for pediatric septic shock. Crit Care Med 2011;39:2511–7.
Wong HR, Wheeler DS, Tegtmeyer K, et al. Toward a clinically feasible gene expression-based subclassification strategy for septic shock: proof of concept. Crit Care Med 2010;38:1955–61.
Standage SW, Caldwell CC, Zingarelli B, Wong HR . Reduced peroxisome proliferator-activated receptor a expression is associated with decreased survival and increased tissue bacterial load in sepsis. Shock 2012;37:164–9.
Eichler GS, Huang S, Ingber DE . Gene Expression Dynamics Inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics 2003;19:2321–2.
Guo Y, Eichler GS, Feng Y, Ingber DE, Huang S . Towards a holistic, yet gene-centered analysis of gene expression profiles: a case study of human lung cancers. J Biomed Biotechnol 2006;2006:69141.
Vanlaere I, Libert C . Matrix metalloproteinases as drug targets in infections caused by gram-negative bacteria and in septic shock. Clin Microbiol Rev 2009;22:224–39, Table of Contents.
Solan PD, Dunsmore KE, Denenberg AG, Odoms K, Zingarelli B, Wong HR . A novel role for matrix metalloproteinase-8 in sepsis. Crit Care Med 2012;40:379–87.
Lauhio A, Hästbacka J, Pettilä V, et al. Serum MMP-8, -9 and TIMP-1 in sepsis: high serum levels of MMP-8 and TIMP-1 are associated with fatal outcome in a multicentre, prospective cohort study. Hypothetical impact of tetracyclines. Pharmacol Res 2011;64:590–4.
Fink MP . Matrix metalloproteinase-8 as a potential drug target for the therapy of sepsis. Crit Care Med 2012;40:655–6.
Prasad AS . Zinc: mechanisms of host defense. J Nutr 2007;137:1345–9.
Heyland DK, Jones N, Cvijanovich NZ, Wong H . Zinc supplementation in critically ill patients: a key pharmaconutrient? JPEN J Parenter Enteral Nutr 2008;32:509–19.
Besecker BY, Exline MC, Hollyfield J, et al. A comparison of zinc metabolism, inflammation, and disease severity in critically ill infected and noninfected adults early after intensive care unit admission. Am J Clin Nutr 2011;93:1356–64.
Cvijanovich NZ, King JC, Flori HR, Gildengorin G, Wong HR . Zinc homeostasis in pediatric critical illness. Pediatr Crit Care Med 2009;10:29–34.
Knoell DL, Liu MJ . Impact of zinc metabolism on innate immune function in the setting of sepsis. Int J Vitam Nutr Res 2010;80:271–7.
Bao S, Liu MJ, Lee B, et al. Zinc modulates the innate immune response in vivo to polymicrobial sepsis through regulation of NF-kappaB. Am J Physiol Lung Cell Mol Physiol 2010;298:L744–54.
Knoell DL, Julian MW, Bao S, et al. Zinc deficiency increases organ damage and mortality in a murine model of polymicrobial sepsis. Crit Care Med 2009;37:1380–8.
Nowak JE, Harmon K, Caldwell CC, Wong HR . Prophylactic zinc supplementation reduces bacterial load and improves survival in a murine model of sepsis. Pediatr Crit Care Med 2012;13:e323–9.
Srinivasan MG, Ndeezi G, Mboijana CK, et al. Zinc adjunct therapy reduces case fatality in severe childhood pneumonia: a randomized double blind placebo-controlled trial. BMC Med 2012;10:14.
Bhatnagar S, Wadhwa N, Aneja S, et al. Zinc as adjunct treatment in infants aged between 7 and 120 days with probable serious bacterial infection: a randomised, double-blind, placebo-controlled trial. Lancet 2012;379:2072–8.
Marshall JC, Reinhart K; International Sepsis Forum. Biomarkers of sepsis. Crit Care Med 2009;37:2290–8.
Kaplan JM, Wong HR . Biomarker discovery and development in pediatric critical care medicine. Pediatr Crit Care Med 2011;12:165–73.
Standage SW, Wong HR . Biomarkers for pediatric sepsis and septic shock. Expert Rev Anti Infect Ther 2011;9:71–9.
Wong HR, Cvijanovich N, Wheeler DS, et al. Interleukin-8 as a stratification tool for interventional trials involving pediatric septic shock. Am J Respir Crit Care Med 2008;178:276–82.
Calfee CS, Thompson BT, Parsons PE, Ware LB, Matthay MA, Wong HR . Plasma interleukin-8 is not an effective risk stratification tool for adults with vasopressor-dependent septic shock. Crit Care Med 2010;38:1436–41.
Wong HR, Salisbury S, Xiao Q, et al. The pediatric sepsis biomarker risk model. Crit Care 2012;16:R174.
Basu RK, Devarajan P, Wong H, Wheeler DS . An update and review of acute kidney injury in pediatrics. Pediatr Crit Care Med 2011;12:339–47.
Wheeler DS, Devarajan P, Ma Q, et al. Serum neutrophil gelatinase-associated lipocalin (NGAL) as a marker of acute kidney injury in critically ill children with septic shock. Crit Care Med 2008;36:1297–303.
Mishra J, Dent C, Tarabishi R, et al. Neutrophil gelatinase-associated lipocalin (NGAL) as a biomarker for acute renal injury after cardiac surgery. Lancet 2005;365:1231–8.
Basu RK, Standage SW, Cvijanovich NZ, et al. Identification of candidate serum biomarkers for severe septic shock-associated kidney injury via microarray. Crit Care 2011;15:R273.
Tang BM, Eslick GD, Craig JC, McLean AS . Accuracy of procalcitonin for sepsis diagnosis in critically ill patients: systematic review and meta-analysis. Lancet Infect Dis 2007;7:210–7.
Wong HR, Cvijanovich NZ, Hall M, et al. Interleukin-27 is a novel candidate diagnostic biomarker for bacterial infection in critically ill children. Crit Care 2012;16:R213.
Wojno ED, Hunter CA . New directions in the basic and translational biology of interleukin-27. Trends Immunol 2012;33:91–7.
Wirtz S, Tubbe I, Galle PR, et al. Protection from lethal septic peritonitis by neutralizing the biological function of interleukin 27. J Exp Med 2006;203:1875–81.
Sun L, Gorospe JR, Hoffman EP, Rao AK . Decreased platelet expression of myosin regulatory light chain polypeptide (MYL9) and other genes with platelet dysfunction and CBFA2/RUNX1 mutation: insights from platelet expression profiling. J Thromb Haemost 2007;5:146–54.
Nowak JE, Wheeler DS, Harmon KK, Wong HR . Admission chemokine (C-C motif) ligand 4 levels predict survival in pediatric septic shock. Pediatr Crit Care Med 2010;11:213–6.
Acknowledgements
The author thanks the following coinvestigators and institutions for submitting biological samples and clinical data for the gene expression database, which could not have been developed without their generous collaboration: Natalie Z. Cvijanovich, Children’s Hospital and Research Center Oakland, Oakland, CA; Mark Hall, Nationwide Children’s Hospital, Columbus, OH; Geoffrey L. Allen, Children’s Mercy Hospital, Kansas City, MO; Neal J. Thomas, Penn State Hershey Children’s Hospital, Hershey, PA; Robert J. Freishtat, Children’s National Medical Center, Washington, DC; Nick Anas, Children’s Hospital of Orange County, Orange, CA; Keith Meyer, Miami Children’s Hospital, Miami, FL; Paul A. Checchia, Texas Children’s Hospital, Houston, TX; Richard Lin, The Children’s Hospital of Philadelphia, Philadelphia, PA; Thomas P. Shanley, CS Mott Children’s Hospital at the University of Michigan, Ann Arbor, MI; Michael T. Bigham, Akron Children’s Hospital, Akron, OH; Anita Sen, Morgan Stanley Children’s Hospital, Columbia University Medical Center, New York, NY; Jeffrey Nowak, Children’s Hospital and Clinics of Minnesota, Minneapolis, MN; Michael Quasney, Children’s Hospital of Wisconsin, Milwaukee, WI; Jared W. Henricksen, Primary Children’s Medical Center, Salt Lake City, UT; and Arun Chopra, St Christopher’s Hospital for Children, Philadelphia, PA.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wong, H. Genome-wide expression profiling in pediatric septic shock. Pediatr Res 73, 564–569 (2013). https://doi.org/10.1038/pr.2013.11
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pr.2013.11
This article is cited by
-
Whole blood transcriptomics identifies subclasses of pediatric septic shock
Critical Care (2023)
-
Transcriptomic data from two primary cell models stimulating human monocytes suggest inhibition of oxidative phosphorylation and mitochondrial function by N. meningitidis which is partially up-regulated by IL-10
BMC Immunology (2017)
-
Differential expression of the Nrf2-linked genes in pediatric septic shock
Critical Care (2015)
-
Differential expression of the nuclear-encoded mitochondrial transcriptome in pediatric septic shock
Critical Care (2014)
-
Systems biology and pediatric research
Pediatric Research (2013)