Abstract
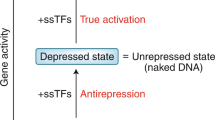
A class of riboswitches that recognizes guanine and discriminates against other purine analogs was recently identified. RNAs that carry the consensus sequence and structural features of guanine riboswitches are located in the 5′ untranslated region (UTR) of numerous prokaryotic genes, where they control the expression of proteins involved in purine salvage and biosynthesis. We report that three representatives of this riboswitch class bind adenine with values for apparent dissociation constant (apparent Kd) that are several orders of magnitude lower than those for binding guanine. Because preference for adenine is attributable to a single nucleotide substitution, the RNA most likely recognizes its ligand by forming a Watson-Crick base pair. In addition, the adenine riboswitch associated with the ydhL gene of Bacillus subtilis functions as a genetic 'on' switch, wherein adenine binding causes a structural rearrangement that precludes formation of an intrinsic transcription terminator stem.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Mandal, M., Boese, B., Winkler, W.C. & Breaker, R.R. Metabolite-sensing riboswitches control fundamental biochemical pathways in bacteria. Cell 113, 577–586 (2003).
Winkler, W.C. & Breaker, R.R. Genetic control by metabolite-binding riboswitches. Chembiochem 4, 1024–1032 (2003).
Sudarsan, N., Wickiser, J.K. Nakamura, S., Ebert, M.S. & Breaker, R.R. An mRNA structure that controls gene expression by binding lysine. Genes Dev. 17, 2685–2697 (2003).
Winkler, W.C., Cohen-Chalamish, S. & Breaker, R.R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. USA 99, 15908–15913 (2002).
Winkler, W.C., Nahvi, A., Sudarsan, N., Barrick, J.E. & Breaker, R.R. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 10, 701–707 (2003).
Nahvi, A. et al. Genetic control by a metabolite binding mRNA. Chem. Biol. 9, 1043–1049 (2002).
Winkler, W., Nahvi, A. & Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956 (2002).
Cristiansen, L.C., Schou, S., Nygaard, P. & Saxild, H.H. Xanthine metabolism in Bacillus subtilis: characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism. J. Bacteriol. 179, 2540–1550 (1997).
Johansen, L.E., Nygaard, P., Lassen, C., Agersø, Y. & Saxild, H.H. Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt-pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL). J. Bacteriol. 185, 5200–5209 (2003).
Soukup, G.A. & Breaker, R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5, 1308–1325 (1999).
Soukup, G.A., DeRose, E.C., Koizumi, M. & Breaker, R.R. Generating new ligand-binding RNAs by affinity maturation and disintegration of allosteric ribozymes. RNA 7, 524–536 (2001).
Peracchi, A., Beigelman, L., Usman, N. & Herschlag, D. Rescue of abasic hammerhead ribozymes by exogenous addition of specific bases. Proc. Natl. Acad. Sci. USA 93, 11522–11527 (1996).
Wilson, K.S. & von Hippel, P.H. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc. Natl. Acad. Sci. USA 92, 8793–8797 (1995).
Gusarov, I & Nudler, E. The mechanism of intrinsic transcription termination. Mol. Cell 4, 495–504 (1999).
Mironov, A.S. et al. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111, 747–756 (2002).
McDaniel, B.A.M., Grundy, F.J., Artsimovitch, I. & Henkin, T.M. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci. USA 100, 3083–3088 (2003).
Epshtein, V., Mironov, A.S. & Nudler, E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl. Acad. Sci. USA 100, 5052–5056 (2003).
Rosenfeld, N., Elowitz, M.B. & Alon, U. Negative autoregulation speeds the response times of transcription networks. J. Mol. Biol. 323, 785–793 (2002).
Grundy, F.J. & Henkin, T.M. The T box and S box transcription termination control systems. Frontiers Biosci. 8, d20–31 (2003).
Grundy, F.J., Winkler, W.C. & Henkin, T.M. tRNA-mediated transcription antitermination in vitro: codon-anticodon pairing independent of the ribosome. Proc. Natl. Acad. Sci. USA 99, 11121–11126 (2002).
Miller, J.H. A Short Course in Bacterial Genetics (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA, 1992).
Acknowledgements
We thank members of the Breaker laboratory for helpful discussions and especially W.C. Winkler for helpful comments on the manuscript. This work was supported by grants from the US National Institutes of Health (GM 559343; NHLBI-N01-HV-28186) and the US National Science Foundation (EIA-0129939; EIA-0323510; EIA-0324045). R.R.B is also grateful for support from the Hereditary Disease Foundation and from the David and Lucile Packard Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Mandal, M., Breaker, R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol 11, 29–35 (2004). https://doi.org/10.1038/nsmb710
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb710
This article is cited by
-
Lithium-sensing riboswitch classes regulate expression of bacterial cation transporter genes
Scientific Reports (2022)
-
Growing a garden of fluorescent RNAs
Nature Chemical Biology (2022)
-
Characterization of Five Purine Riboswitches in Cellular and Cell-Free Expression Systems
Current Microbiology (2022)
-
Unraveling RNA dynamical behavior of TPP riboswitches: a comparison between Escherichia coli and Arabidopsis thaliana
Scientific Reports (2019)
-
Life times of metastable states guide regulatory signaling in transcriptional riboswitches
Nature Communications (2018)