Abstract
CRISPR adaptive immune systems protect bacteria from infections by deploying CRISPR RNA (crRNA)-guided enzymes to recognize and cut foreign nucleic acids. Type VI-A CRISPR–Cas systems include the Cas13a enzyme, an RNA-activated RNase capable of crRNA processing and single-stranded RNA degradation upon target-transcript binding. Here we present the 2.0-Å resolution crystal structure of a crRNA-bound Lachnospiraceae bacterium Cas13a (LbaCas13a), representing a recently discovered Cas13a enzyme subtype. This structure and accompanying biochemical experiments define the Cas13a catalytic residues that are directly responsible for crRNA maturation. In addition, the orientation of the foreign-derived target-RNA-specifying sequence in the protein interior explains the conformational gating of Cas13a nuclease activation. These results describe how Cas13a enzymes generate functional crRNAs and how catalytic activity is blocked before target-RNA recognition, with implications for both bacterial immunity and diagnostic applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dy, R.L., Richter, C., Salmond, G.P. & Fineran, P.C. Remarkable mechanisms in microbes to resist phage infections. Annu. Rev. Virol. 1, 307–331 (2014).
Barrangou, R. & Marraffini, L.A. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell 54, 234–244 (2014).
Marraffini, L.A. CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61 (2015).
Hsu, P.D., Lander, E.S. & Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014).
Wright, A.V., Nuñez, J.K. & Doudna, J.A. Biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering. Cell 164, 29–44 (2016).
Barrangou, R. & Doudna, J.A. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 34, 933–941 (2016).
O'Connell, M.R. et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516, 263–266 (2014).
Shmakov, S. et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell 60, 385–397 (2015).
Nelles, D.A. et al. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell 165, 488–496 (2016).
East-Seletsky, A. et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016).
Shmakov, S. et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 15, 169–182 (2017).
Smargon, A.A. et al. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell 65, 618–630.e7 (2017).
Gootenberg, J.S. et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442 (2017).
East-Seletsky, A., O′Connell, M.R., Burstein, D., Knott, G.J. & Doudna, J.A. RNA targeting by functionally orthogonal type VI-A CRISPR-Cas enzymes. Mol. Cell 66, 373–383.e3 (2017).
Mojica, F.J., Díez-Villaseñor, C., García-Martínez, J. & Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740 (2009).
Liu, L. et al. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell 168, 121–134.e12 (2017).
Abudayyeh, O.O. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016).
Haurwitz, R.E., Jinek, M., Wiedenheft, B., Zhou, K. & Doudna, J.A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329, 1355–1358 (2010).
Sternberg, S.H., Haurwitz, R.E. & Doudna, J.A. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA 18, 661–672 (2012).
Li, H. Structural principles of CRISPR RNA processing. Structure 23, 13–20 (2015).
Hochstrasser, M.L. & Doudna, J.A. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem. Sci. 40, 58–66 (2015).
Charpentier, E., Richter, H., van der Oost, J. & White, M.F. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol. Rev. 39, 428–441 (2015).
Schirle, N.T., Sheu-Gruttadauria, J. & MacRae, I.J. Structural basis for microRNA targeting. Science 346, 608–613 (2014).
Liu, L. et al. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell 170, 714–726.e10 (2017).
Jiang, F. & Doudna, J.A. The structural biology of CRISPR-Cas systems. Curr. Opin. Struct. Biol. 30, 100–111 (2015).
Nakanishi, K. Anatomy of RISC: how do small RNAs and chaperones activate Argonaute proteins? Wiley Interdiscip. Rev. RNA 7, 637–660 (2016).
Osawa, T., Inanaga, H., Sato, C. & Numata, T. Crystal structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog. Mol. Cell 58, 418–430 (2015).
Parker, J.S., Parizotto, E.A., Wang, M., Roe, S.M. & Barford, D. Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Mol. Cell 33, 204–214 (2009).
Jiang, F., Zhou, K., Ma, L., Gressel, S. & Doudna, J.A. STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science 348, 1477–1481 (2015).
Salomon, W.E., Jolly, S.M., Moore, M.J., Zamore, P.D. & Serebrov, V. Single-molecule imaging reveals that Argonaute reshapes the binding properties of its nucleic acid guides. Cell 162, 84–95 (2015).
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Winn, M.D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Evans, P.R. & Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013).
Powell, H.R. The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 55, 1690–1695 (1999).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Terwilliger, T.C. et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Keating, K.S. & Pyle, A.M. Semiautomated model building for RNA crystallography using a directed rotameric approach. Proc. Natl. Acad. Sci. USA 107, 8177–8182 (2010).
Afonine, P.V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012).
Terwilliger, T.C. et al. Iterative-build OMIT maps: map improvement by iterative model building and refinement without model bias. Acta Crystallogr. D Biol. Crystallogr. 64, 515–524 (2008).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Acknowledgements
We thank N. Ma and K. Zhou for technical assistance, C.L. Gee for assistance with refinement, the 8.3.1 beamline staff at the Advanced Light Source, and members of the Doudna laboratory for helpful discussions. G.J.K. acknowledges support from the Howard Hughes Medical Institute. This work was supported in part by the Paul Allen Institute, the National Science Foundation (grant MCB-1244557), and the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
G.J.K., A.E.-S., M.R.O., and J.A.D. designed the study. G.J.K., A.E-S., J.C.C., and E.C. expressed and purified proteins. G.J.K. prepared and crystallized the Cas13a complexes with assistance from J.C.C., collected X-ray data, and determined the crystal structure with support from J.M.H. A.E-S. carried out biochemical assays with assistance from G.J.K. and J.C.C. G.J.K. drafted the manuscript with assistance from A.E.-S., and all authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The Regents of the University of California have patents pending for CRISPR technologies on which the authors are inventors.
Integrated supplementary information
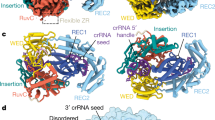
Supplementary Figure 1 Anomalous scattering within the asymmetric unit of LbaCa13a, RNA content of crystals, and crRNA electron density maps.
a, Opposing perspectives of LbaCas13a are shown as a transparent cartoon with domains colored as in Fig. 1c. The anomalously scattering iodides, sulfurs, and phosphates are shown as yellow, orange, and blue colored spheres, respectively. The anomalous Fourier difference map shown is contoured to 3σ. b, Urea-PAGE gel of 20-nt and 24-nt spacer-containing pre-crRNA substrates incubated with LbaCas13a to produce the complex prepared for crystallization. Crystals of each complex were looped, washed, and dissolved to determine the presence of full-length mature crRNA after crystallization. c, Simulated annealing mFo-DFc omit electron density map of the entire crRNA over two orthogonal views contoured to 2σ. d, The discontinuous crRNA contained within the asymmetric unit (two copies denoted as 1 and 2) of the 20-nt spacer-containing structure of LbaCas13a. Simulated annealing mFo-DFc omit electron density map of the entire crRNA contoured to 3σ. e, The discontinuous crRNA contained within the 28-nt spacer containing structure of LshCas13a (Liu et al., Cell, 168, 121-134, 2017). Simulated annealing mFo-DFc omit electron density map of the entire crRNA contoured to 3σ.
Supplementary Figure 2 Structural comparison of LbaCas13a and LshCas13a.
a. Side-by-side comparison of LbaCas13a (left) and LshCas13a (right). The structures are shown as cartoons with domains colored as in Fig. 1, with domain boundaries indicated schematically beneath the structures. b-h, Structural alignment of each individual domain of LbaCas13a (colored) to that of LshCas13a (grey). i, Structural alignment of the LbaCas13a NTD (teal) with the LshCas13a NTD (grey), using the crRNA repeat as a reference point for superposition. j, Structural alignment of the LbaCas13a crRNA (orange) and LshCas13a crRNA (grey).
Supplementary Figure 3 Schematic representation of contacts between LbaCas13a and crRNA.
a, The contact map illustrates only those contacts that have unambiguous electron density. Nucleotides that make up the 5′ handle and 3′ spacer are labelled as orange or blue, respectively. Planar stacking interactions are indicated by dashed lines, and hydrogen bonds/electrostatic interactions are indicated by arrows. Amino acids that are conserved within the A-cleaving subfamily or across the entire Cas13a family are indicated with one or two asterisks, respectively. b-c, Conserved LbaCas13a amino acid contacts to the sugar-phosphate backbone of spacer nucleotides A(6) and A(8).
Supplementary Figure 4 Biochemical measurements of crRNA affinity and dual catalytic activity of LbaCas13a.
a, Filter-binding assays measuring the affinity of LbaCas13a for its cognate crRNA. The quantified binding data was fit to a standard binding isotherm with measured dissociation constants (mean ± st. dev., n= 3) of 0.98 ± 0.08 nM for a cognate crRNA and 84.7 ± 12.6 nM for a representative off target ssRNA. b-c, The dependence of LbaCas13a ssRNA targeting activation on spacer sequence length (20-28 nt) was measured by fluorescent cleavage assays. Reactions were carried out as described in the Methods, except that cleavage buffer was used instead of processing buffer. (b) Apparent rates (mean ± st. dev., n= 3) were fitted to resulting time courses and (c) endpoint fluorescent values (background corrected mean ± st. dev., n= 3) were measured. All crRNA with various spacer lengths were able to direct LbaCas13a for ssRNA cleavage, although the shortest spacer tested (20-nt) reached a lower plateau value while retaining a similar apparent rate compared to all other spacer lengths. d, pre-crRNA processing assays were performed as described in the Methods for alanine substitutions of residues interacting with the 5′ end of the mature crRNA in the LbaCas13a binary structure. Quantified data were fit to single-exponential decays with calculated pseudo-first order rate constants (kobs) (mean ± st. dev., n= 3) as follows: Lba Wildtype (WT), 0.109 ± 0.005 min−1; Lba W325A, 0.011 ± 0.004 min−1; and Lba N1232A, 0.014 ± 0.005 min−1. Rates could not be calculated for H328A, K432A, K435A, K1305A, and K1320A. Endpoint data from these curves is presented in Fig 4. e-f, ssRNA targeting by LbaCas13a and alanine substitution mutations was carried out consistent with b-c. Asterisks mark mutants for which rates could not be fit. Binding affinity of LbaCas13a mutants to (g) pre-crRNA dG(-29) (pre-crRNA mimic) or (h) mature crRNA substrates measured by filter-binding. Measured binding affinities were fit to a standard binding isotherm and are summarized in (i). j, Representative gel of pre-crRNA processing by LbaCas13a in the presence and absence of divalent metal ion chelator, EDTA for 60 mins. k, pre-crRNA processing by LbaCas13a carried out as described in the Methods for a pre-crRNA substrate bearing 8- or 2-nt 5′ to the scissile phosphate.
Supplementary Figure 5 Multiple-sequence amino acid alignment of Cas13a homologs.
Multiple sequence alignment of entire Cas13a family adapted from (East-Seletsky et al., Mol Cell, 66, 373-383, 2017). The local regions in the Cas13a 2 Helical-1 domain (a) and HEPN2 domain (b) implicated in pre-crRNA processing across LbaCas12a (this study), LbuCas13a and LshCas13a (Liu et al., Cell, 168, 121-134, 2017) noted with a symbol above or below the mutated residue. Residues are marked according to the functional result of their mutation: processing deficiency (red triangles), minimal impact (yellow squares,) and no effect (teal diamonds). Symbols below the LshCas13a sequence correspond to mutations made to LshCas13a by1. Coloration of the alignment denotes residue conservation according to the ClustalX scheme, in which darker hues indicate stronger conservation.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 (PDF 1599 kb)
Rights and permissions
About this article
Cite this article
Knott, G., East-Seletsky, A., Cofsky, J. et al. Guide-bound structures of an RNA-targeting A-cleaving CRISPR–Cas13a enzyme. Nat Struct Mol Biol 24, 825–833 (2017). https://doi.org/10.1038/nsmb.3466
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3466
This article is cited by
-
Orthogonal inducible control of Cas13 circuits enables programmable RNA regulation in mammalian cells
Nature Communications (2024)
-
dCas13-mediated translational repression for accurate gene silencing in mammalian cells
Nature Communications (2024)
-
CRISPR/Cas genome editing in plants: mechanisms, applications, and overcoming bottlenecks
Functional & Integrative Genomics (2024)
-
CRISPR–Cas13d in plant biology: an insight
Plant Biotechnology Reports (2024)
-
Development of Cas13a-based therapy for cancer treatment
Molecular Biology Reports (2024)