Abstract
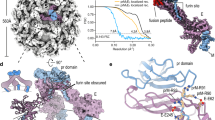
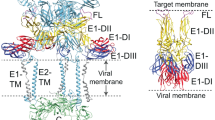
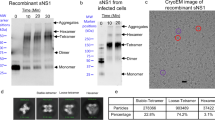
Regulated by pH, membrane-anchored proteins E and M function during dengue virus maturation and membrane fusion. Our atomic model of the whole virion from cryo–electron microscopy at 3.5-Å resolution reveals that in the mature virus at neutral extracellular pH, the N-terminal 20-amino-acid segment of M (involving three pH-sensing histidines) latches and thereby prevents spring-loaded E fusion protein from prematurely exposing its fusion peptide. This M latch is fastened at an earlier stage, during maturation at acidic pH in the trans-Golgi network. At a later stage, to initiate infection in response to acidic pH in the late endosome, M releases the latch and exposes the fusion peptide. Thus, M serves as a multistep chaperone of E to control the conformational changes accompanying maturation and infection. These pH-sensitive interactions could serve as targets for drug discovery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
WHO. Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control (WHO, Geneva, Switzerland, 2009).
Borio, L. et al. Hemorrhagic fever viruses as biological weapons: medical and public health management. J. Am. Med. Assoc. 287, 2391–2405 (2002).
Yu, I.M. et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319, 1834–1837 (2008).
Li, L. et al. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science 319, 1830–1834 (2008).
Kielian, M. & Rey, F.A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4, 67–76 (2006).
Harrison, S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 (2008).
Kuhn, R.J. et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108, 717–725 (2002).
Zhang, W. et al. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10, 907–912 (2003).
Modis, Y., Ogata, S., Clements, D. & Harrison, S.C. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 100, 6986–6991 (2003).
Modis, Y., Ogata, S., Clements, D. & Harrison, S.C. Structure of the dengue virus envelope protein after membrane fusion. Nature 427, 313–319 (2004).
Zhang, Y. et al. Conformational changes of the flavivirus E glycoprotein. Structure 12, 1607–1618 (2004).
Plevka, P. et al. Maturation of flaviviruses starts from one or more icosahedrally independent nucleation centres. EMBO Rep. 12, 602–606 (2011).
Liu, X., Jiang, W., Jakana, J. & Chiu, W. Averaging tens to hundreds of icosahedral particle images to resolve protein secondary structure elements using a Multi-Path Simulated Annealing optimization algorithm. J. Struct. Biol. 160, 11–27 (2007).
Rosenthal, P.B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Kaptein, S.J. et al. A derivate of the antibiotic doxorubicin is a selective inhibitor of dengue and yellow fever virus replication in vitro. Antimicrob. Agents Chemother. 54, 5269–5280 (2010).
Wang, Q.Y. et al. A small-molecule dengue virus entry inhibitor. Antimicrob. Agents Chemother. 53, 1823–1831 (2009).
Cockburn, J.J. et al. Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature 432, 122–125 (2004).
Zhang, R. et al. 4.4 A cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J. 30, 3854–3863 (2011).
Fritz, R. et al. The unique transmembrane hairpin of flavivirus fusion protein E is essential for membrane fusion. J. Virol. 85, 4377–4385 (2011).
Fritz, R., Stiasny, K. & Heinz, F.X. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J. Cell Biol. 183, 353–361 (2008).
Kroschewski, H., Sagripanti, J.L. & Davidson, A.D. Identification of amino acids in the dengue virus type 2 envelope glycoprotein critical to virus infectivity. J. Gen. Virol. 90, 2457–2461 (2009).
Nelson, S., Poddar, S., Lin, T.-Y. & Pierson, T.C. Protonation of individual histidine residues is not required for the pH-dependent entry of West Nile Virus: evaluation of the “histidine switch” hypothesis. J. Virol. 83, 12631–12635 (2009).
Yu, I.M. et al. Association of the pr peptides with dengue virus at acidic pH blocks membrane fusion. J. Virol. 83, 12101–12107 (2009).
Zhang, Y. et al. Structures of immature flavivirus particles. EMBO J. 22, 2604–2613 (2003).
Gaudin, Y. Reversibility in fusion protein conformational changes. The intriguing case of rhabdovirus-induced membrane fusion. Subcell. Biochem. 34, 379–408 (2000).
Lalezari, J.P. et al. A phase II clinical study of the long-term safety and antiviral activity of enfuvirtide-based antiretroviral therapy. AIDS 17, 691–698 (2003).
Ludtke, S.J., Baldwin, P.R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999).
Liang, Y., Ke, E.Y. & Zhou, Z.H. IMIRS: a high-resolution 3D reconstruction package integrated with a relational image database. J. Struct. Biol. 137, 292–304 (2002).
Liu, H. et al. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329, 1038–1043 (2010).
Mindell, J.A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Kivioja, T., Ravantti, J., Verkhovsky, A., Ukkonen, E. & Bamford, D. Local average intensity-based method for identifying spherical particles in electron micrographs. J. Struct. Biol. 131, 126–134 (2000).
Zhang, J. et al. Mechanism of folding chamber closure in a group II chaperonin. Nature 463, 379–383 (2010).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Li, Y. & Zhang, Y. REMO: A new protocol to refine full atomic protein models from C-alpha traces by optimizing hydrogen-bonding networks. Proteins 76, 665–676 (2009).
Brunger, A.T. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2, 2728–2733 (2007).
Ge, P. & Zhou, Z.H. Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. Proc. Natl. Acad. Sci. USA 108, 9637–9642 (2011).
Acknowledgements
We thank V. Vordam (Centers for Disease Control Dengue Branch, San Juan, Puerto Rico) for providing the viral stock and advising about cell culture, I. Atanasov and W.H. Hui for participation in data acquisition, J. Jiang for suggestions in data processing, UCLA undergraduate students K.M. Lau, J. Chen and K. Chen and B.K. Zhou of Beverly Vista School for scanning photographic films and boxing particles, and A. Paredes and J.-Q. Zhang for preliminary efforts in viral preparation. This work is supported in part by grants from the US National Institutes of Health grant GM071940 (to Z.H.Z.), National Natural Science Foundation of China (NSFC) grant 30928003 and 30725017 (to G.B.), NSFC 30470085 and 30870480 (to Q.Z.). We acknowledge the use of instruments at the Electron Imaging Center for NanoMachines supported by National Institutes of Health (1S10RR23057) and the California NanoSystems Institute at UCLA.
Author information
Authors and Affiliations
Contributions
Z.H.Z., X.Z., P.G. and X.Y. designed experiments. J.M.B., X.Z. and X.Y. cultured cells and purified virus samples. X.Z., X.Y., P.G., J.M.B. and Z.H.Z. obtained cryo-EM images. Z.H.Z., J.M.B. and X.Z. participated in the image processing and three-dimensional reconstruction from the Polara data. X.Z. obtained a 7-Å structure from the Polara data. P.G. refined the structure to 3.5-Å resolution with the Titan Krios data and built the atomic models. P.G., X.Z. and Z.H.Z. interpreted the structure and drafted the manuscript. P.G., X.Z., Z.H.Z. and S.S. finalized the manuscript. G.B. and Q.Z. participated in discussion and interpretation of the results. All authors reviewed the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Tables 1 and 2 (PDF 4306 kb)
Supplementary Movie 1
A 3D visualization of various structures described in the figures. The animation begins with a surface rendering of the cryoEM density map, rotating around a 2-fold axis. Structural units containing membrane proteins E and M shown in the same color are equivalent by icosahedral symmetry. The differently colored structural units are quasiequivalent. Specifically, the green units fall on the icosahedral 5-fold axes, the blue on the 3-fold and the red on the 2-fold. This scene is followed by a close up view of a rhombus-shaped group of six E-M dimers, fitted with the ribbon representations of its atomic model, rotating around the horizontal axis. Next, the three quasi-equivalent E-M-M-E heterotetramers are averaged. Rotated around the horizontal axis, half of this averaged tetramer is rendered as a shaded surface representation and half is shown in semi-transparent grey, superimposed with the ribbon representations of an E monomer and an M monomer, with the same color scheme as in Figure 2. (AVI 26509 kb)
Supplementary Movie 2
Interactions between E (molecular surface) and M (sticks). First, an animated view of Figure 4b. Second, an animated view of the molecular surface of E and the ribbon and stick model of M that comprise pocket 1, as shown in Figure 4c and Supplementary Figure 8c. A second pocket 1 in a symmetry related position is also visible in the same view. Third, an animated view of the molecular surface of E and the ribbon and stick model of M that comprise pocket 2, as shown Figure 4d and Supplementary Figure 8e. The histidine in the center of this movie is His7 of M which is involved in the pH sensitive latching of E by M. The two contiguous nitrogen atoms in a nearby bulge above and to the right of this His7 belong to His209 of E. Finally, an animated view of the molecular surface of E and the ribbon and stick model of M that comprise pocket 3 as shown Figure 4e and Supplementary Figure 8g. The surrounding of the central Trp19 of M is highly hydrophobic as indicated by the atom types on the molecular surface of E. (AVI 12159 kb)
Rights and permissions
About this article
Cite this article
Zhang, X., Ge, P., Yu, X. et al. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat Struct Mol Biol 20, 105–110 (2013). https://doi.org/10.1038/nsmb.2463
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2463
This article is cited by
-
Identification of the flavivirus conserved residues in the envelope protein hinge region for the rational design of a candidate West Nile live-attenuated vaccine
npj Vaccines (2023)
-
Zika virus M protein latches and locks the E protein from transitioning to an immature state after prM cleavage
npj Viruses (2023)
-
Adaptation to host cell environment during experimental evolution of Zika virus
Communications Biology (2022)
-
Designer DNA nanostructures for viral inhibition
Nature Protocols (2022)
-
Evolution and activation mechanism of the flavivirus class II membrane-fusion machinery
Nature Communications (2022)