Abstract
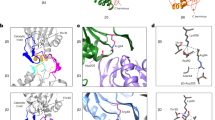
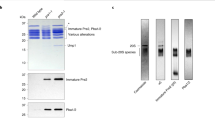
The Saccharomyces cerevisiae proteasome comprises a 19-subunit regulatory particle and a 28-subunit core particle. To be degraded, substrates must cross the core particle–regulatory particle interface, a site for complex conformational changes and regulatory events. This interface includes two aligned heteromeric rings, one formed by the six ATPase (Rpt) subunits of the regulatory particle and the other by the seven α subunits of the core particle. The Rpt C termini bind to intersubunit cavities in the α-ring, thus directing core particle gating and proteasome assembly. We mapped the Rpt C termini to the α subunit pockets, using a cross-linking approach that revealed an unexpected asymmetry: one side of the ring shows 1:1 contacts of Rpt2-α4, Rpt6-α3 and Rpt3-α2, whereas on the opposite side, the Rpt1, Rpt4 and Rpt5 tails each cross-link to multiple α pockets. Rpt–core particle cross-links are all sensitive to nucleotides, implying that ATP hydrolysis drives dynamic alterations at the core particle–regulatory particle interface.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 (2009).
Schrader, E.K., Harstad, K.G. & Matouschek, A. Targeting proteins for degradation. Nat. Chem. Biol. 5, 815–822 (2009).
Demartino, G.N. & Gillette, T.G. Proteasomes: machines for all reasons. Cell 129, 659–662 (2007).
Groll, M. et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 386, 463–471 (1997).
Yu, Y. et al. Interactions of PAN's C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions. EMBO J. 29, 692–702 (2010).
Stadtmueller, B.M. et al. Structural models for interactions between the 20S proteasome and its PAN/19S activators. J. Biol. Chem. 285, 13–17 (2010).
Rabl, J. et al. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 30, 360–368 (2008).
Smith, D.M. et al. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol. Cell 27, 731–744 (2007).
Förster, A., Masters, E.I., Whitby, F.G., Robinson, H. & Hill, C.P. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol. Cell 18, 589–599 (2005).
Köhler, A. et al. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol. Cell 7, 1143–1152 (2001).
Whitby, F.G. et al. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature 408, 115–120 (2000).
Groll, M. et al. A gated channel into the proteasome core particle. Nat. Struct. Biol. 7, 1062–1067 (2000).
Bajorek, M., Finley, D. & Glickman, M.H. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr. Biol. 13, 1140–1144 (2003).
Smith, D.M., Fraga, H., Reis, C., Kafri, G. & Goldberg, A.L. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell 144, 526–538 (2011).
Zhang, F. et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell 34, 473–484 (2009).
Djuranovic, S. et al. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol. Cell 34, 580–590 (2009).
Smith, D.M. et al. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol. Cell 20, 687–698 (2005).
Glickman, M.H. et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94, 615–623 (1998).
Weber-Ban, E.U., Reid, B.G., Miranker, A.D. & Horwich, A.L. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature 401, 90–93 (1999).
Sauer, R.T. & Baker, T.A. AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80, 587–612 (2011).
Glynn, S.E., Martin, A., Nager, A.R., Baker, T.A. & Sauer, R.T. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell 139, 744–756 (2009).
Aubin-Tam, M.E., Olivares, A.O., Sauer, R.T., Baker, T.A. & Lang, M.J. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell 145, 257–267 (2011).
Maillard, R.A. et al. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell 145, 459–469 (2011).
Saeki, Y., Toh, E.A., Kudo, T., Kawamura, H. & Tanaka, K. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell 137, 900–913 (2009).
Kaneko, T. et al. Assembly pathway of the mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell 137, 914–925 (2009).
Funakoshi, M., Tomko, R.J. Jr., Kobayashi, H. & Hochstrasser, M. Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell 137, 887–899 (2009).
Gillette, T.G., Kumar, B., Thompson, D., Slaughter, C.A. & DeMartino, G.N. Differential roles of the COOH termini of AAA subunits of PA700 (19S regulator) in asymmetric assembly and activation of the 26S proteasome. J. Biol. Chem. 283, 31813–31822 (2008).
Roelofs, J. et al. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature 459, 861–865 (2009).
Park, S. et al. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature 459, 866–870 (2009).
Kumar, B., Kim, Y.C. & DeMartino, G.N. The C terminus of Rpt3, an ATPase subunit of PA700 (19 S) regulatory complex, is essential for 26 S proteasome assembly but not for activation. J. Biol. Chem. 285, 39523–39535 (2010).
Thompson, D., Hakala, K. & DeMartino, G.N. Subcomplexes of PA700, the 19 S regulator of the 26 S proteasome, reveal relative roles of AAA subunits in 26 S proteasome assembly and activation and ATPase activity. J. Biol. Chem. 284, 24891–24903 (2009).
Kusmierczyk, A.R., Kunjappu, M.J., Funakoshi, M. & Hochstrasser, M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat. Struct. Mol. Biol. 15, 237–244 (2008).
Bohn, S. et al. Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc. Natl. Acad. Sci. USA 107, 20992–20997 (2010).
Nickell, S. et al. Insights into the molecular architecture of the 26S proteasome. Proc. Natl. Acad. Sci. USA 106, 11943–11947 (2009).
da Fonseca, P.C. & Morris, E.P. Structure of the human 26S proteasome: subunit radial displacements open the gate into the proteolytic core. J. Biol. Chem. 283, 23305–23314 (2008).
Andréasson, C., Fiaux, J., Rampelt, H., Druffel-Augustin, S. & Bukau, B. Insights into the structural dynamics of the Hsp110-Hsp70 interaction reveal the mechanism for nucleotide exchange activity. Proc. Natl. Acad. Sci. USA 105, 16519–16524 (2008).
Chen, L.L., Rosa, J.J. Turner, S. & Pepinsky, R.B. Production of multimeric forms of CD4 through a sugar-based cross-linking strategy. J. Biol. Chem. 266, 18237–18243 (1991).
Tomko, R.J. Jr., Funakoshi, M., Schneider, K., Wang, J. & Hochstrasser, M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Mol. Cell 38, 393–403 (2010).
Martin, A., Baker, T.A. & Sauer, R.T. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat. Struct. Mol. Biol. 15, 1147–1151 (2008).
Wang, J. et al. Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure 9, 177–184 (2001).
Bochtler, M. et al. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature 403, 800–805 (2000).
Kleijnen, M.F. et al. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat. Struct. Mol. Biol. 14, 1180–1188 (2007).
Chen, C. et al. Subunit-subunit interactions in the human 26S proteasome. Proteomics 8, 508–520 (2008).
Satoh, K., Sasajima, H., Nyoumura, K.I., Yokosawa, H. & Sawada, H. Assembly of the 26S proteasome is regulated by phosphorylation of the p45/Rpt6 ATPase subunit. Biochemistry 40, 314–319 (2001).
Hartmann-Petersen, R., Tanaka, K. & Hendil, K.B. Quaternary structure of the ATPase complex of human 26S proteasomes determined by chemical cross-linking. Arch. Biochem. Biophys. 386, 89–94 (2001).
Davy, A. et al. A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep. 2, 821–828 (2001).
Zhang, Z. et al. Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J. Biol. Chem. 275, 15157–15165 (2000).
Gerlinger, U.M., Guckel, R., Hoffmann, M., Wolf, D.H. & Hilt, W. Yeast cycloheximide-resistant crl mutants are proteasome mutants defective in protein degradation. Mol. Biol. Cell 8, 2487–2499 (1997).
Walz, J. et al. 26S proteasome structure revealed by three-dimensional electron microscopy. J. Struct. Biol. 121, 19–29 (1998).
Park, S., Tian, G., Roelofs, J. & Finley, D. Assembly manual for the proteasome regulatory particle: the first draft. Biochem. Soc. Trans. 38, 6–13 (2010).
Leggett, D.S. et al. Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 (2002).
Enemark, E.J. & Joshua-Tor, L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442, 270–275 (2006).
Thomsen, N.D. & Berger, J.M. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell 139, 523–534 (2009).
Davies, J.M., Brunger, A.T. & Weis, W.I. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure 16, 715–726 (2008).
Rose, M.D., Winston, F.M. & Heiter, P. Methods in Yeast Genetics: A Laboratory Course Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1990).
Finley, D., Ozkaynak, E. & Varshavsky, A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48, 1035–1046 (1987).
Janke, C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004).
Schmidt, M. et al. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat. Struct. Mol. Biol. 12, 294–303 (2005).
Leggett, D.S., Glickman, M.H. & Finley, D. Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol. Biol. 301, 57–70 (2005).
Elsasser, S., Schmidt, M. & Finley, D. Characterization of the proteasome using native gel electrophoresis. Methods Enzymol. 398, 353–363 (2005).
Acknowledgements
We thank W. Tansey (Vanderbilt University Medical Center) and C. Mann (Commissariat à l'énergie atomique et aux énergies alternatives (CEA)/Saclay) for antibodies, B.-H. Lee for helpful discussions, A. Matouschek for comments on the manuscript and W. Baumeister for permission to reproduce Figure 6D. Funding was provided by US National Institutes of Health grants to D.F. (R37GM43601), C.P.H. (R01 GM59135) and S.G. (GM67945). S.P. was supported by a fellowship from the Charles A. King Trust and M.J.L. by the American Health Assistance Foundation.
Author information
Authors and Affiliations
Contributions
G.T., S.P., C.P.H. and D.F. contributed to the conception of this project. G.T., S.P. and B.H. contributed to strain construction and genetic analysis. G.T, S.P. and M.J.L. conducted cross-linking studies. F.M. and S.P.G. carried out mass spectrometry on cross-linked samples. G.T., C.P.H. and D.F. were largely responsible for the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 1 and 2 (PDF 4817 kb)
Rights and permissions
About this article
Cite this article
Tian, G., Park, S., Lee, M. et al. An asymmetric interface between the regulatory and core particles of the proteasome. Nat Struct Mol Biol 18, 1259–1267 (2011). https://doi.org/10.1038/nsmb.2147
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2147
This article is cited by
-
Minimal mechanistic component of HbYX-dependent proteasome activation that reverses impairment by neurodegenerative-associated oligomers
Communications Biology (2023)
-
An atomic structure of the human 26S proteasome
Nature Structural & Molecular Biology (2016)
-
Phosphorylation of the C-terminal tail of proteasome subunit α7 is required for binding of the proteasome quality control factor Ecm29
Scientific Reports (2016)
-
Proteasome Activation is Mediated via a Functional Switch of the Rpt6 C-terminal Tail Following Chaperone-dependent Assembly
Scientific Reports (2015)
-
ATP binding to neighbouring subunits and intersubunit allosteric coupling underlie proteasomal ATPase function
Nature Communications (2015)