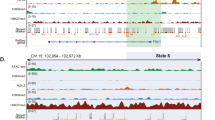
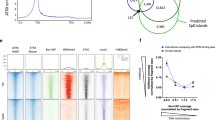
Abstract
Binding to nucleosomal DNA is critical for 'pioneer' transcription factors such as the winged-helix transcription factors Foxa1 and Foxa2 to regulate chromatin structure and gene activation. Here we report the genome-wide map of nucleosome positions in the mouse liver, with emphasis on transcriptional start sites, CpG islands, Foxa2 binding sites and their correlation with gene expression. Despite the heterogeneity of liver tissue, we could clearly discern the nucleosome pattern of the predominant liver cell, the hepatocyte. By analyzing nucleosome occupancy and the distributions of heterochromatin protein 1 (Hp1), CBP (also known as Crebbp) and p300 (Ep300) in Foxa1- and Foxa2-deficient livers, we find that the maintenance of nucleosome position and chromatin structure surrounding Foxa2 binding sites is independent of Foxa1 and Foxa2.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Schones, D.E. et al. Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887–898 (2008).
Shivaswamy, S. et al. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 6, e65 (2008).
Valouev, A. et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 18, 1051–1063 (2008).
Beato, M. & Eisfeld, K. Transcription factor access to chromatin. Nucleic Acids Res. 25, 3559–3563 (1997).
McPherson, C.E., Shim, E.Y., Friedman, D.S. & Zaret, K.S. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell 75, 387–398 (1993).
Chen, H., Li, B. & Workman, J.L. A histone-binding protein, nucleoplasmin, stimulates transcription factor binding to nucleosomes and factor-induced nucleosome disassembly. EMBO J. 13, 380–390 (1994).
Li, B., Adams, C.C. & Workman, J.L. Nucleosome binding by the constitutive transcription factor Sp1. J. Biol. Chem. 269, 7756–7763 (1994).
Kaestner, K.H., Hiemisch, H. & Schutz, G. Targeted disruption of the gene encoding hepatocyte nuclear factor 3γ results in reduced transcription of hepatocyte-specific genes. Mol. Cell. Biol. 18, 4245–4251 (1998).
Shen, W., Scearce, L.M., Brestelli, J.E., Sund, N.J. & Kaestner, K.H. Foxa3 (hepatocyte nuclear factor 3γ) is required for the regulation of hepatic GLUT2 expression and the maintenance of glucose homeostasis during a prolonged fast. J. Biol. Chem. 276, 42812–42817 (2001).
Lee, C.S., Friedman, J.R., Fulmer, J.T. & Kaestner, K.H. The initiation of liver development is dependent on Foxa transcription factors. Nature 435, 944–947 (2005).
Li, Z. et al. Foxa1 and Foxa2 regulate bile duct development in mice. J. Clin. Invest. 119, 1537–1545 (2009).
Zaret, K. Developmental competence of the gut endoderm: genetic potentiation by GATA and HNF3/fork head proteins. Dev. Biol. 209, 1–10 (1999).
Hoffman, B.G. et al. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Res. 20, 1037–1051 (2010).
Gardiner-Garden, M. & Frommer, M. CpG islands in vertebrate genomes. J. Mol. Biol. 196, 261–282 (1987).
Kundu, T.K. & Rao, M.R. CpG islands in chromatin organization and gene expression. J. Biochem. 125, 217–222 (1999).
Yaragatti, M., Basilico, C. & Dailey, L. Identification of active transcriptional regulatory modules by the functional assay of DNA from nucleosome-free regions. Genome Res. 18, 930–938 (2008).
Lai, E., Prezioso, V.R., Tao, W.F., Chen, W.S. & Darnell, J.E. Jr. Hepatocyte nuclear factor 3 α belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 5, 416–427 (1991).
Tuteja, G., Jensen, S.T., White, P. & Kaestner, K.H. Cis-regulatory modules in the mammalian liver: composition depends on strength of Foxa2 consensus site. Nucleic Acids Res. 36, 4149–4157 (2008).
Lupien, M. et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132, 958–970 (2008).
Xu, J. et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 23, 2824–2838 (2009).
Chodavarapu, R.K. et al. Relationship between nucleosome positioning and DNA methylation. Nature 466, 388–392 (2010).
Sutton, J. et al. Genesis, a winged helix transcriptional repressor with expression restricted to embryonic stem cells. J. Biol. Chem. 271, 23126–23133 (1996).
Fleischmann, G., Filipski, R. & Elgin, S.C. Isolation and distribution of a Drosophila protein preferentially associated with inactive regions of the genome. Chromosoma 96, 83–90 (1987).
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001).
Visel, A. et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 (2009).
Heintzman, N.D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007).
Greenbaum, L.E. et al. CCAAT enhancer–binding protein β is required for normal hepatocyte proliferation in mice after partial hepatectomy. J. Clin. Invest. 102, 996–1007 (1998).
Morrison, A.J., Sardet, C. & Herrera, R.E. Retinoblastoma protein transcriptional repression through histone deacetylation of a single nucleosome. Mol. Cell. Biol. 22, 856–865 (2002).
Rubins, N.E. et al. Transcriptional networks in the liver: hepatocyte nuclear factor 6 function is largely independent of Foxa2. Mol. Cell. Biol. 25, 7069–7077 (2005).
Tuteja, G., White, P., Schug, J. & Kaestner, K.H. Extracting transcription factor targets from ChIP-Seq data. Nucleic Acids Res. 37, e113 (2009).
Crooks, G.E., Hon, G., Chandonia, J.M. & Brenner, S.E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Gao, N. et al. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 22, 3435–3448 (2008).
Tusher, V.G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116–5121 (2001).
Acknowledgements
We thank A. Fox, O. Smirnova, K. Brondell, A. Chen, A. Riblett and J. LaRossa (University of Pennsylvania) for excellent technical support. This study was supported by the US National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (P01-DK049210 to K.H.K.). Z.L. was supported by National Sciences and Engineering Research Council of Canada and Juvenile Diabetes Research Foundation postdoctoral fellowship awards. We would like to acknowledge support of the Institute for Diabetes, Obesity, and Metabolism Functional Genomics Core by an NIDDK Research Center grant (P30DK19525).
Author information
Authors and Affiliations
Contributions
Z.L. did the majority of experiments and computational analyses. J.S., P.W. and G.T. did a part of the experiments and computational analyses. K.H.K. directed the whole study. Z.L. and K.H.K. designed the experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 (PDF 1380 kb)
Rights and permissions
About this article
Cite this article
Li, Z., Schug, J., Tuteja, G. et al. The nucleosome map of the mammalian liver. Nat Struct Mol Biol 18, 742–746 (2011). https://doi.org/10.1038/nsmb.2060
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2060
This article is cited by
-
Role of cell-type specific nucleosome positioning in inducible activation of mammalian promoters
Nature Communications (2020)
-
NUCLIZE for quantifying epigenome: generating histone modification data at single-nucleosome resolution using genuine nucleosome positions
BMC Genomics (2019)
-
Knockdown of FOXA2 enhances the osteogenic differentiation of bone marrow-derived mesenchymal stem cells partly via activation of the ERK signalling pathway
Cell Death & Disease (2018)
-
Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders
Genome Biology (2016)
-
Nucleosome architecture throughout the cell cycle
Scientific Reports (2016)