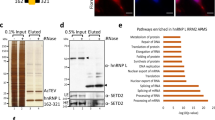
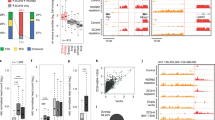
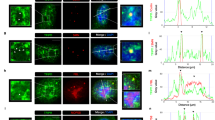
Abstract
Pre-messenger RNAs (pre-mRNAs) maturation is initiated cotranscriptionally. It is therefore conceivable that chromatin-borne information participates in alternative splicing. Here we find that elevated levels of trimethylation of histone H3 on Lys9 (H3K9me3) are a characteristic of the alternative exons of several genes including CD44. On this gene the chromodomain protein HP1γ, frequently defined as a transcriptional repressor, facilitates inclusion of the alternative exons via a mechanism involving decreased RNA polymerase II elongation rate. In addition, accumulation of HP1γ on the variant region of the CD44 gene stabilizes association of the pre-mRNA with the chromatin. Altogether, our data provide evidence for localized histone modifications impacting alternative splicing. They further implicate HP1γ as a possible bridging molecule between the chromatin and the maturating mRNA, with a general impact on splicing decisions.
This is a preview of subscription content, access via your institution
Access options







Similar content being viewed by others
Accession codes
References
Wang, E.T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008).
Allemand, E., Batsche, E. & Muchardt, C. Splicing, transcription, and chromatin: a menage a trois. Curr. Opin. Genet. Dev. 18, 145–151 (2008).
Auboeuf, D. et al. Differential recruitment of nuclear receptor coactivators may determine alternative RNA splice site choice in target genes. Proc. Natl. Acad. Sci. USA 101, 2270–2274 (2004).
Singh, J. & Padgett, R.A. Rates of in situ transcription and splicing in large human genes. Nat. Struct. Mol. Biol. 16, 1128–1133 (2009).
Listerman, I., Sapra, A.K. & Neugebauer, K.M. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 13, 815–822 (2006).
Das, R. et al. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell 26, 867–881 (2007).
Hargreaves, D.C., Horng, T. & Medzhitov, R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138, 129–145 (2009).
Batsché, E., Yaniv, M. & Muchardt, C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 13, 22–29 (2006).
Kornblihtt, A.R., Schor, I.E., Allo, M. & Blencowe, B.J. When chromatin meets splicing. Nat. Struct. Mol. Biol. 16, 902–903 (2009).
Schwartz, S., Meshorer, E. & Ast, G. Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 16, 990–995 (2009).
Tilgner, H. et al. Nucleosome positioning as a determinant of exon recognition. Nat. Struct. Mol. Biol. 16, 996–1001 (2009).
Andersson, R., Enroth, S., Rada-Iglesias, A., Wadelius, C. & Komorowski, J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 19, 1732–1741 (2009).
Brehm, A., Tufteland, K.R., Aasland, R. & Becker, P.B. The many colours of chromodomains. Bioessays 26, 133–140 (2004).
Mujtaba, S., Zeng, L. & Zhou, M.M. Structure and acetyl-lysine recognition of the bromodomain. Oncogene 26, 5521–5527 (2007).
Ruthenburg, A.J., Li, H., Patel, D.J. & Allis, C.D. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 (2007).
Schor, I.E., Rascovan, N., Pelisch, F., Allo, M. & Kornblihtt, A.R. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc. Natl. Acad. Sci. USA 106, 4325–4330 (2009).
Sims, R.J. III et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell 28, 665–676 (2007).
Kolasinska-Zwierz, P. et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 41, 376–381 (2009).
Hon, G., Wang, W. & Ren, B. Discovery and annotation of functional chromatin signatures in the human genome. PLOS Comput. Biol. 5, e1000566 (2009).
Luco, R.F. et al. Regulation of alternative splicing by histone modifications. Science 327, 996–1000 (2010).
Alló, M. et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 16, 717–724 (2009).
Tyagi, A., Ryme, J., Brodin, D., Ostlund Farrants, A.K. & Visa, N. SWI/SNF associates with nascent pre-mRNPs and regulates alternative pre-mRNA processing. PLoS Genet. 5, e1000470 (2009).
de la Mata, M. et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12, 525–532 (2003).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Vakoc, C.R., Mandat, S.A., Olenchock, B.A. & Blobel, G.A. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell 19, 381–391 (2005).
Mateescu, B., Bourachot, B., Rachez, C., Ogryzko, V. & Muchardt, C. Regulation of an inducible promoter by an HP1beta-HP1gamma switch. EMBO Rep. 9, 267–272 (2008).
Smallwood, A., Esteve, P.O., Pradhan, S. & Carey, M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 21, 1169–1178 (2007).
König, H., Ponta, H. & Herrlich, P. Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J. 17, 2904–2913 (1998).
Lomberk, G., Bensi, D., Fernandez-Zapico, M.E. & Urrutia, R. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat. Cell Biol. 8, 407–415 (2006).
Newman, A.J. & Nagai, K. Structural studies of the spliceosome: blind men and an elephant. Curr. Opin. Struct. Biol. 20, 82–89 (2010).
du Chéné, I. et al. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 26, 424–435 (2007).
Rowe, H.M. et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463, 237–240 (2010).
Stewart, M.D., Li, J. & Wong, J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol. Cell. Biol. 25, 2525–2538 (2005).
Fritsch, L. et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol. Cell 37, 46–56 (2010).
Muchardt, C. et al. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 3, 975–981 (2002).
Cannistra, S.A., DeFranzo, B., Niloff, J. & Ottensmeir, C. Functional heterogeneity of CD44 molecules in ovarian cancer cell lines. Clin. Cancer Res. 1, 333–342 (1995).
Elgadi, K.M., Meguid, R.A., Qian, M., Souba, W.W. & Abcouwer, S.F. Cloning and analysis of unique human glutaminase isoforms generated by tissue-specific alternative splicing. Physiol. Genomics 1, 51–62 (1999).
Vicent, G.P. et al. Induction of progesterone target genes requires activation of erk and msk kinases and phosphorylation of histone H3. Mol. Cell 24, 367–381 (2006).
Lavigne, M. et al. Interaction of HP1 and Brg1/Brm with the globular domain of histone H3 is required for HP1-mediated repression. PLoS Genet. 5, e1000769 (2009).
Flanagin, S., Nelson, J.D., Castner, D.G., Denisenko, O. & Bomsztyk, K. Microplate-based chromatin immunoprecipitation method, Matrix ChIP: a platform to study signaling of complex genomic events. Nucleic Acids Res. 36, e17 (2008).
Loomis, R.J. et al. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol. Cell 33, 450–461 (2009).
de Wit, E., Greil, F. & van Steensel, B. High-resolution mapping reveals links of HP1 with active and inactive chromatin components. PLoS Genet. 3, e38. doi:10.1371/journal.pgen.0030038.
Piacentini, L. et al. Heterochromatin protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila. PLoS Genet. 5, e1000670 (2009).
Piacentini, L., Fanti, L., Berloco, M., Perrini, B. & Pimpinelli, S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J. Cell Biol. 161, 707–714 (2003).
Mikkelsen, T.S. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007).
Brinkman, A.B. et al. Histone modification patterns associated with the human X chromosome. EMBO Rep. 7, 628–634 (2006).
Tachibana, M. et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3–K9. Genes Dev. 19, 815–826 (2005).
Verdel, A. et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303, 672–676 (2004).
Moazed, D. Small RNAs in transcriptional gene silencing and genome defence. Nature 457, 413–420 (2009).
Melcher, M. et al. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol. Cell. Biol. 20, 3728–3741 (2000).
Sampath, S.C. et al. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol. Cell 27, 596–608 (2007).
Matter, N., Herrlich, P. & Konig, H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420, 691–695 (2002).
Bonnet, F., Vigneron, M., Bensaude, O. & Dubois, M.F. Transcription-independent phosphorylation of the RNA polymerase II C-terminal domain (CTD) involves ERK kinases (MEK1/2). Nucleic Acids Res. 27, 4399–4404 (1999).
Wahl, M.C., Will, C.L. & Luhrmann, R. The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718 (2009).
Morgenstern, J.P. & Land, H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18, 3587–3596 (1990).
Méndez, J. & Stillman, B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602–8612 (2000).
de la Grange, P., Gratadou, L., Delord, M., Dutertre, M. & Auboeuf, D. Splicing factor and exon profiling across human tissues. Nucleic Acids Res. 38, 2825–2838 (2010).
Acknowledgements
We thank E. Allemand and J. Seeler for critical reading of the manuscript, and D. Auboeuf (INSERM U590, Centre Léon Bérard, France), P de la Grange (Genosplice Technology, France), V. Ogryzko (CNRS, IGR, Université Paris-XI, France), L. Fritsch and S. Ait-Si-Ali (Université Paris–Diderot, Paris, France) for advice and gifts of reagents. V.S.-A. received fellowships from Région Ile-de-France and L'Association pour la Recherche sur le Cancer. The work was supported by grants from the Agence National de la Recherche and Cancéropôle Ile-de-France.
Author information
Authors and Affiliations
Contributions
V.S.-A., E.B. and C.R. designed, performed and analyzed the experiments and prepared the manuscript. C.M. conceived the project and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 1 and 2 (PDF 555 kb)
Rights and permissions
About this article
Cite this article
Saint-André, V., Batsché, E., Rachez, C. et al. Histone H3 lysine 9 trimethylation and HP1γ favor inclusion of alternative exons. Nat Struct Mol Biol 18, 337–344 (2011). https://doi.org/10.1038/nsmb.1995
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1995
This article is cited by
-
DNA topoisomerase 1 represses HIV-1 promoter activity through its interaction with a guanine quadruplex present in the LTR sequence
Retrovirology (2023)
-
Functions of HP1 proteins in transcriptional regulation
Epigenetics & Chromatin (2022)
-
The Heterochromatin protein 1 is a regulator in RNA splicing precision deficient in ulcerative colitis
Nature Communications (2022)
-
TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control
Nature Communications (2020)
-
Citrullination of HP1γ chromodomain affects association with chromatin
Epigenetics & Chromatin (2019)