Abstract
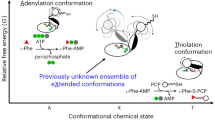
The metazoan cytosolic fatty acid synthase (FAS) contains all of the enzymes required for de novo fatty acid biosynthesis covalently linked around two reaction chambers. Although the three-dimensional architecture of FAS has been mostly defined, it is unclear how reaction intermediates can transfer between distant catalytic domains. Using single-particle EM, we have identified a near continuum of conformations consistent with a remarkable flexibility of FAS. The distribution of conformations was influenced by the presence of substrates and altered by different catalytic mutations, suggesting a direct correlation between conformation and specific enzymatic activities. We interpreted three-dimensional reconstructions by docking high-resolution structures of individual domains, and they show that the substrate-loading and condensation domains dramatically swing and swivel to access substrates within either reaction chamber. Concomitant rearrangement of the β-carbon–processing domains synchronizes acyl chain reduction in one chamber with acyl chain elongation in the other.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Sul, H.S. & Smith, S. Fatty acid synthesis in eukaryotes. in Biochemistry of Lipids, Lipoproteins and Membranes (ed. Vance, D.E.a.V. J.E.) 155–190 (Elsevier, Amsterdam; Oxford, 2008).
Kuhajda, F.P. et al. Fatty acid synthesis: a potential selective target for antineoplastic therapy. Proc. Natl. Acad. Sci. USA 91, 6379–6383 (1994).
Loftus, T.M. et al. Reduced food intake and body weight in mice treated with fatty acid synthase inhibitors. Science 288, 2379–2381 (2000).
Jenni, S. et al. Structure of fungal fatty acid synthase and implications for iterative substrate shuttling. Science 316, 254–261 (2007).
Lomakin, I.B., Xiong, Y. & Steitz, T.A. The crystal structure of yeast fatty acid synthase, a cellular machine with eight active sites working together. Cell 129, 319–332 (2007).
Asturias, F.J. et al. Structure and molecular organization of mammalian fatty acid synthase. Nat. Struct. Mol. Biol. 12, 225–232 (2005).
Maier, T., Jenni, S. & Ban, N. Architecture of mammalian fatty acid synthase at 4.5 Å resolution. Science 311, 1258–1262 (2006).
Rangan, V.S., Joshi, A.K. & Smith, S. Mapping the functional topology of the animal fatty acid synthase by mutant complementation in vitro. Biochemistry 40, 10792–10799 (2001).
Witkowski, A. et al. Dibromopropanone cross-linking of the phosphopantetheine and active-site cysteine thiols of the animal fatty acid synthase can occur both inter- and intrasubunit. Reevaluation of the side-by-side, antiparallel subunit model. J. Biol. Chem. 274, 11557–11563 (1999).
Burgess, S.A., Walker, M.L., Thirumurugan, K., Trinick, J. & Knight, P.J. Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J. Struct. Biol. 147, 247–258 (2004).
Joshi, A.K., Witkowski, A., Berman, H.A., Zhang, L. & Smith, S. Effect of modification of the length and flexibility of the acyl carrier protein-thioesterase interdomain linker on functionality of the animal fatty acid synthase. Biochemistry 44, 4100–4107 (2005).
Radermacher, M. The three-dimensional reconstruction of single particles from random and non-random tilt series. J. Electron Microsc. Tech. 9, 359–394 (1988).
Keatinge-Clay, A.T. & Stroud, R.M. The structure of a ketoreductase determines the organization of the β-carbon processing enzymes of modular polyketide synthases. Structure 14, 737–748 (2006).
Smith, S. & Tsai, S.C. The type I fatty acid and polyketide synthases: a tale of two megasynthases. Nat. Prod. Rep. 24, 1041–1072 (2007).
Horton, J.R., Sawada, K., Nishibori, M. & Cheng, X. Structural basis for inhibition of histamine N-methyltransferase by diverse drugs. J. Mol. Biol. 353, 334–344 (2005).
Tang, Y., Kim, C.Y., Mathews, I.I., Cane, D.E. & Khosla, C. The 2.7-angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc. Natl. Acad. Sci. USA 103, 11124–11129 (2006).
Cheng, Y. et al. Single particle reconstructions of the transferrin-transferrin receptor complex obtained with different specimen preparation techniques. J. Mol. Biol. 355, 1048–1065 (2006).
Joshi, A.K. & Smith, S. Construction, expression, and characterization of a mutated animal fatty acid synthase deficient in the dehydrase function. J. Biol. Chem. 268, 22508–22513 (1993).
Witkowski, A., Joshi, A.K., Lindqvist, Y. & Smith, S. Conversion of a β-ketoacyl synthase to a malonyl decarboxylase by replacement of the active-site cysteine with glutamine. Biochemistry 38, 11643–11650 (1999).
Tang, Y., Chen, A.Y., Kim, C.Y., Cane, D.E. & Khosla, C. Structural and mechanistic analysis of protein interactions in module 3 of the 6-deoxyerythronolide B synthase. Chem. Biol. 14, 931–943 (2007).
Chen, Z.J. et al. Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new insights into its substrate recognition properties. J. Mol. Biol. 379, 830–844 (2008).
Joshi, A.K., Rangan, V.S., Witkowski, A. & Smith, S. Engineering of an active animal fatty acid synthase dimer with only one competent subunit. Chem. Biol. 10, 169–173 (2003).
Maier, T., Leibundgut, M. & Ban, N. The crystal structure of a mammalian fatty acid synthase. Science 321, 1315–1322 (2008).
Smith, S. & Abraham, S. Fatty acid synthase from lactating rat mammary gland. Methods Enzymol. 35, 65–74 (1975).
Tischendorf, G.W., Zeichhardt, H. & Stoffler, G. Determination of the location of proteins L14, L17, L18, L19, L22, L23 on the surface of the 50S ribosomal subunit of Escherichia coli by immune electron microscopy. Mol. Gen. Genet. 134, 187–208 (1974).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
Rath, B.K. & Frank, J. Fast automatic particle picking from cryo-electron micrographs using a locally normalized cross-correlation function: a case study. J. Struct. Biol. 145, 84–90 (2004).
Penczek, P., Radermacher, M. & Frank, J. Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy 40, 33–53 (1992).
Bretaudiere, J.P. & Frank, J. Reconstitution of molecule images analysed by correspondence analysis: a tool for structural interpretation. J. Microsc. 144, 1–14 (1986).
Radermacher, M., Wagenknecht, T., Verschoor, A. & Frank, J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J. Microsc. 146, 113–136 (1987).
Keatinge-Clay, A.T. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem. Biol. 14, 898–908 (2007).
Pasta, S., Witkowski, A., Joshi, A.K. & Smith, S. Catalytic residues are shared between two pseudosubunits of the dehydratase domain of the animal fatty acid synthase. Chem. Biol. 14, 1377–1385 (2007).
Soding, J., Biegert, A. & Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Ploskon, E. et al. A mammalian type I fatty acid synthase acyl carrier protein domain does not sequester acyl chains. J. Biol. Chem. 283, 518–528 (2008).
Volkmann, N. & Hanein, D. Quantitative fitting of atomic models into observed densities derived by electron microscopy. J. Struct. Biol. 125, 176–184 (1999).
Koski, M.K., Haapalainen, A.M., Hiltunen, J.K. & Glumoff, T. A two-domain structure of one subunit explains unique features of eukaryotic hydratase 2. J. Biol. Chem. 279, 24666–24672 (2004).
Oefner, C., Schulz, H., D'Arcy, A. & Dale, G.E. Mapping the active site of Escherichia coli malonyl-CoA-acyl carrier protein transacylase (FabD) by protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 62, 613–618 (2006).
Bunkoczi, G. et al. Mechanism and substrate recognition of human holo ACP synthase. Chem. Biol. 14, 1243–1253 (2007).
Acknowledgements
We thank A. Witkowski for helpful discussions. We also acknowledge the National Resource for Automated Molecular Microscopy for assistance with data collection. The work was supported by a research fellowship F32 DK080622 (to E.J.B.) and grant RO1 DK16073 (to S.S.) from the US National Institutes of Health.
Author information
Authors and Affiliations
Contributions
E.J.B. performed all experiments and data analysis; S.S. provided purified FAS; all authors contributed to designing experiments, interpreting results and writing the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 (PDF 2633 kb)
Supplementary Video 1
2D analysis reveals a continuum of domain rearrangements within the β-carbon processing portion of FAS. (GIF 2693 kb)
Supplementary Video 2
An animation illustrates how FAS may interchange between the experimentally observed conformations to facilitate contacts between the ACP and active sites of the catalytic domains in a typical reaction cycle. (MOV 9889 kb)
Rights and permissions
About this article
Cite this article
Brignole, E., Smith, S. & Asturias, F. Conformational flexibility of metazoan fatty acid synthase enables catalysis. Nat Struct Mol Biol 16, 190–197 (2009). https://doi.org/10.1038/nsmb.1532
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1532
This article is cited by
-
Direct structural analysis of a single acyl carrier protein domain in fatty acid synthase from the fungus Saccharomyces cerevisiae
Communications Biology (2024)
-
Atomic model for core modifying region of human fatty acid synthase in complex with Denifanstat
Nature Communications (2023)
-
Enzymology of assembly line synthesis by modular polyketide synthases
Nature Chemical Biology (2023)
-
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis
BMC Biology (2022)
-
Porous metal-metalloporphyrin gel as catalytic binding pocket for highly efficient synergistic catalysis
Nature Communications (2019)