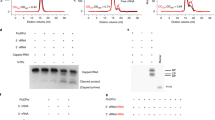
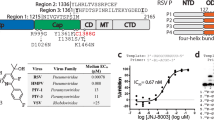
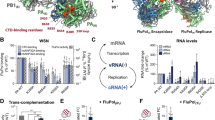
Abstract
Influenza virus mRNAs are synthesized by the trimeric viral polymerase using short capped primers obtained by a 'cap-snatching' mechanism. The polymerase PB2 subunit binds the 5′ cap of host pre-mRNAs, which are cleaved after 10–13 nucleotides by the PB1 subunit. Using a library-screening method, we identified an independently folded domain of PB2 that has specific cap binding activity. The X-ray structure of the domain with bound cap analog m7GTP at 2.3-Å resolution reveals a previously unknown fold and a mode of ligand binding that is similar to, but distinct from, other cap binding proteins. Binding and functional studies with point mutants confirm that the identified site is essential for cap binding in vitro and cap-dependent transcription in vivo by the trimeric polymerase complex. These findings clarify the nature of the cap binding site in PB2 and will allow efficient structure-based design of new anti-influenza compounds inhibiting viral transcription.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Olsen, B. et al. Global patterns of influenza a virus in wild birds. Science 312, 384–388 (2006).
Horimoto, T. & Kawaoka, Y. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 3, 591–600 (2005).
Elton, D., Digard, P., Tiley, L. & Ortín, J. Structure and function of the influenza virus RNP. in Current Topics in Influenza Virology (ed. Kawaoka, Y.) (Horizon Scientific, Norfolk, 2005).
Plotch, S.J., Bouloy, M., Ulmanen, I. & Krug, R.M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23, 847–858 (1981).
Li, M.L., Rao, P. & Krug, R.M. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 20, 2078–2086 (2001).
Poon, L.L., Pritlove, D.C., Fodor, E. & Brownlee, G.G. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J. Virol. 73, 3473–3476 (1999).
Cianci, C., Tiley, L. & Krystal, M. Differential activation of the influenza virus polymerase via template RNA binding. J. Virol. 69, 3995–3999 (1995).
Honda, A., Mizumoto, K. & Ishihama, A. Two separate sequences of PB2 subunit constitute the RNA cap-binding site of influenza virus RNA polymerase. Genes Cells 4, 475–485 (1999).
Fechter, P. et al. Two aromatic residues in the PB2 subunit of influenza A RNA polymerase are crucial for cap binding. J. Biol. Chem. 278, 20381–20388 (2003).
Fechter, P. & Brownlee, G.G. Recognition of mRNA cap structures by viral and cellular proteins. J. Gen. Virol. 86, 1239–1249 (2005).
Area, E. et al. 3D structure of the influenza virus polymerase complex: localization of subunit domains. Proc. Natl. Acad. Sci. USA 101, 308–313 (2004).
Torreira, E. et al. Three-dimensional model for the isolated recombinant influenza virus polymerase heterotrimer. Nucleic Acids Res. 35, 3774–3783 (2007).
Tarendeau, F. et al. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat. Struct. Mol. Biol. 14, 229–233 (2007).
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 (1993).
Marcotrigiano, J., Gingras, A.C., Sonenberg, N. & Burley, S.K. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89, 951–961 (1997).
Mazza, C., Segref, A., Mattaj, I.W. & Cusack, S. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 21, 5548–5557 (2002).
Hodel, A.E., Gershon, P.D. & Quiocho, F.A. Structural basis for sequence-nonspecific recognition of 5′-capped mRNA by a cap-modifying enzyme. Mol. Cell 1, 443–447 (1998).
Niedzwiecka, A. et al. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E–BP1 proteins. J. Mol. Biol. 319, 615–635 (2002).
Martin-Benito, J. et al. Three-dimensional reconstruction of a recombinant influenza virus ribonucleoprotein particle. EMBO Rep. 2, 313–317 (2001).
Ortega, J. et al. Ultrastructural and functional analyses of recombinant influenza virus ribonucleoproteins suggest dimerization of nucleoprotein during virus amplification. J. Virol. 74, 156–163 (2000).
Worch, R. et al. Specificity of recognition of mRNA 5′ cap by human nuclear cap-binding complex. RNA 11, 1355–1363 (2005).
Hooker, L. et al. Quantitative analysis of influenza virus RNP interaction with RNA cap structures and comparison to human cap binding protein eIF4E. Biochemistry 42, 6234–6240 (2003).
Bouloy, M., Plotch, S.J. & Krug, R.M. Both the 7-methyl and the 2′-O-methyl groups in the cap of mRNA strongly influence its ability to act as primer for influenza virus RNA transcription. Proc. Natl. Acad. Sci. USA 77, 3952–3956 (1980).
Fodor, E. et al. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76, 8989–9001 (2002).
Hara, K., Schmidt, F.I., Crow, M. & Brownlee, G.G. Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J. Virol. 80, 7789–7798 (2006).
Penn, C.R. & Mahy, B.W. Capped mRNAs may stimulate the influenza virion polymerase by allosteric modulation. Virus Res. 1, 1–13 (1984).
Kawakami, K., Mizumoto, K., Ishihama, A., Shinozaki-Yamaguchi, K. & Miura, K. Activation of influenza virus-associated RNA polymerase by cap-1 structure (m7GpppNm). J. Biochem. 97, 655–661 (1985).
Cianci, C., Colonno, R.J. & Krystal, M. Differential effect of modified capped RNA substrates on influenza virus transcription. Virus Res. 50, 65–75 (1997).
Li, M.L., Ramirez, B.C. & Krug, R.M. RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J. 17, 5844–5852 (1998).
Ulmanen, I., Broni, B. & Krug, R.M. Influenza virus temperature-sensitive cap (m7GpppNm)-dependent endonuclease. J. Virol. 45, 27–35 (1983).
Yamanaka, K. et al. Characterization of a temperature-sensitive mutant in the RNA polymerase PB2 subunit gene of influenza A/WSN/33 virus. Arch. Virol. 114, 65–73 (1990).
Luo, G., Danetz, S. & Krystal, M. Inhibition of influenza viral polymerases by minimal viral RNA decoys. J. Gen. Virol. 78, 2329–2333 (1997).
De Clercq, E. Antiviral agents active against influenza A viruses. Nat. Rev. Drug Discov. 5, 1015–1025 (2006).
DuBridge, R.B. et al. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7, 379–387 (1987).
Ortin, J., Najera, R., Lopez, C., Davila, M. & Domingo, E. Genetic variability of Hong Kong (H3N2) influenza viruses: spontaneous mutations and their location in the viral genome. Gene 11, 319–331 (1980).
Falcon, A.M. et al. Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein. J. Virol. 78, 3880–3888 (2004).
Neumann, G. et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96, 9345–9350 (1999).
de la Luna, S., Martinez, C. & Ortin, J. Molecular cloning and sequencing of influenza virus A/Victoria/3/75 polymerase genes: sequence evolution and prediction of possible functional domains. Virus Res. 13, 143–155 (1989).
Van Duyne, G.D., Standaert, R.F., Karplus, P.A., Schreiber, S.L. & Clardy, J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993).
Perrakis, A., Morris, R. & Lamzin, V.S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 (1999).
Murshudov, G.N. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Lovell, S.C. et al. Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins 50, 437–450 (2003).
Barcena, J. et al. Monoclonal antibodies against influenza virus PB2 and NP polypeptides interfere with the initiation step of viral mRNA synthesis in vitro. J. Virol. 68, 6900–6909 (1994).
Perales, B., de la Luna, S., Palacios, I. & Ortin, J. Mutational analysis identifies functional domains in the influenza A virus PB2 polymerase subunit. J. Virol. 70, 1678–1686 (1996).
Acknowledgements
We thank P. Mas for help with ESPRIT screening; H. Belrhali, A. McCarthy and M. Walsh (MRC-France) for help in data collection and structure determination; Y. Fernández (CSIC) for help in the polymerase assays; and B. Dublet, L. Signor and E. Forest (IBS) for MS measurements. We acknowledge the ESRF, EMBL and MRC-France for access to synchrotron facilities, the Partnership for Structural Biology for an integrated structural biology environment and the EU for funding of the FLUPOL contract (SP5B-CT-2007-044263). Work at CNB was also supported by the Spanish Ministry of Education and Science (Ministerio de Educación y Ciencia) (BFU2004-491) and the Research Programme VIRHOST-CM from Comunidad de Madrid (S-SAL/0185/2006). Thanks to Catherine for inspiration.
Author information
Authors and Affiliations
Contributions
D.J.H. and F.T. devised and implemented the extension of ESPRIT to internal domains and identified soluble central fragments of PB2. D.G. and F.T. purified and biochemically characterized soluble central fragments of PB2. D.G. identified and crystallized the minimal cap binding domain, made and crystallized selenomethionated protein and made and assayed cap binding of domain mutants. S.C. performed the crystal structure analysis with T.C. P.R.-I. established conditions for binding of polymerase to cap-analog resins. P.R.-I. and R.C. contributed equally to the generation of and functional characterization of mutants in the context of recombinant polymerase and RNPs. J.O. designed and supervised the mutant phenotype analyses. S.C. compiled the text with contributions from J.O. and D.H. P.S. and J.L. performed and analysed the SPR measurements. J.O., R.W.H.R. and S.C. are principal investigators of the EU FP6 FLUPOL project.
Corresponding author
Ethics declarations
Competing interests
D.J.H. and F.T. have applied for a patent on ESPRIT technology. S.C., D.G., D.J.H. and F.T. have applied for a patent on the use of recombinantly expressed constructs of the PB2 cap binding domain and the three-dimensional structure for anti-influenza drug design.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1 and 2 and Supplementary Methods (PDF 3271 kb)
Rights and permissions
About this article
Cite this article
Guilligay, D., Tarendeau, F., Resa-Infante, P. et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat Struct Mol Biol 15, 500–506 (2008). https://doi.org/10.1038/nsmb.1421
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1421
This article is cited by
-
Phosphorylation of the PA subunit of influenza polymerase at Y393 prevents binding of the 5′-termini of RNA and polymerase function
Scientific Reports (2023)
-
Cap-snatching inhibitors of influenza virus are inhibitory to the in vitro transcription of rice stripe virus
Phytopathology Research (2022)
-
An overview of influenza A virus genes, protein functions, and replication cycle highlighting important updates
Virus Genes (2022)
-
A novel E198K substitution in the PA gene of influenza A virus with reduced susceptibility to baloxavir acid
Archives of Virology (2022)
-
Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity
Nature Communications (2021)