Abstract
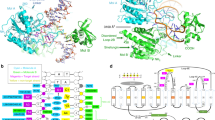
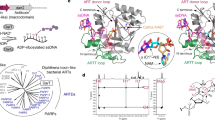
DNA-adenine methylation at certain GATC sites plays a pivotal role in bacterial and phage gene expression as well as bacterial virulence. We report here the crystal structures of the bacteriophage T4Dam DNA adenine methyltransferase (MTase) in a binary complex with the methyl-donor product S-adenosyl-L-homocysteine (AdoHcy) and in a ternary complex with a synthetic 12-bp DNA duplex and AdoHcy. T4Dam contains two domains: a seven-stranded catalytic domain that harbors the binding site for AdoHcy and a DNA binding domain consisting of a five-helix bundle and a β-hairpin that is conserved in the family of GATC-related MTase orthologs. Unexpectedly, the sequence-specific T4Dam bound to DNA in a nonspecific mode that contained two Dam monomers per synthetic duplex, even though the DNA contains a single GATC site. The ternary structure provides a rare snapshot of an enzyme poised for linear diffusion along the DNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lacks, S. & Greenberg, B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J. Mol. Biol. 114, 153–168 (1977).
Hattman, S., Brooks, J.E. & Masurekar, M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J. Mol. Biol. 126, 367–380 (1978).
Bale, A., d'Alarcao, M. & Marinus, M.G. Characterization of DNA adenine methylation mutants of Escherichia coli K12. Mutat. Res. 59, 157–165 (1979).
Julio, S.M. et al. DNA adenine methylase is essential for viability and plays a role in the pathogenesis of Yersinia pseudotuberculosis and Vibrio cholerae. Infect. Immun. 69, 7610–7615 (2001).
Garcia-Del Portillo, F., Pucciarelli, M.G. & Casadesus, J. DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc. Natl. Acad. Sci. USA 96, 11578–11583 (1999).
Heithoff, D.M., Sinsheimer, R.L., Low, D.A. & Mahan, M.J. An essential role for DNA adenine methylation in bacterial virulence. Science 284, 967–970 (1999).
Heithoff, D.M. et al. Salmonella DNA adenine methylase mutants confer cross-protective immunity. Infect. Immun. 69, 6725–6730 (2001).
Dueger, E.L., House, J.K., Heithoff, D.M. & Mahan, M.J. Salmonella DNA adenine methylase mutants elicit protective immune responses to homologous and heterologous serovars in chickens. Infect. Immun. 69, 7950–7954 (2001).
Dueger, E.L., House, J.K., Heithoff, D.M. & Mahan, M.J. Salmonella DNA adenine methylase mutants prevent colonization of newly hatched chickens by homologous and heterologous serovars. Int. J. Food Microbiol. 80, 153–159 (2003).
Urieli-Shoval, S., Gruenbaum, Y. & Razin, A. Sequence and substrate specificity of isolated DNA methylases from Escherichia coli C. J. Bacteriol. 153, 274–280 (1983).
Messer, W. & Noyer-Weidner, M. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell 54, 735–737 (1988).
Modrich, P. Methyl-directed DNA mismatch correction. J. Biol. Chem. 264, 6597–6600 (1989).
Yang, W. Structure and function of mismatch repair proteins. Mutat. Res. 460, 245–256 (2000).
Lu, M., Campbell, J.L., Boye, E. & Kleckner, N. SeqA: a negative modulator of replication initiation in E. coli. Cell 77, 413–426 (1994).
Kang, S., Lee, H., Han, J.S. & Hwang, D.S. Interaction of SeqA and Dam methylase on the hemimethylated origin of Escherichia coli chromosomal DNA replication. J. Biol. Chem. 274, 11463–11468 (1999).
Guarne, A., Zhao, Q., Ghirlando, R. & Yang, W. Insights into negative modulation of E. coli replication initiation from the structure of SeqA-hemimethylated DNA complex. Nat. Struct. Biol. 9, 839–843 (2002).
Oshima, T. et al. Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol. Microbiol. 45, 673–695 (2002).
Løbner-Olesen, A., Marinus, M.G. & Hansen, F.G. Role of SeqA and Dam in Escherichia coli gene expression: a global/microarray analysis. Proc. Natl. Acad. Sci. USA 100, 4672–4677 (2003).
Julio, S.M., Heithoff, D.M., Sinsheimer, R.L., Low, D.A. & Mahan, M.J. DNA adenine methylase overproduction in Yersinia pseudotuberculosis alters YopE expression and secretion and host immune responses to infection. Infect. Immun. 70, 1006–1009 (2002).
Hattman, S. Unusual transcriptional and translational regulation of the bacteriophage Mu mom operon. Pharmacol. Ther. 84, 367–388 (1999).
Malone, T., Blumenthal, R.M. & Cheng, X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 253, 618–632 (1995).
Klimasauskas, S., Kumar, S., Roberts, R.J. & Cheng, X. HhaI methyltransferase flips its target base out of the DNA helix. Cell 76, 357–369 (1994).
Goedecke, K., Pignot, M., Goody, R.S., Scheidig, A.J. & Weinhold, E. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nat. Struct. Biol. 8, 121–125 (2001).
Reinisch, K.M., Chen, L., Verdine, G.L. & Lipscomb, W.N. The crystal structure of HaeIII methyltransferase covalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell 82, 143–153 (1995).
Gong, W., O'Gara, M., Blumenthal, R.M. & Cheng, X. Structure of PvuII DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res. 25, 2702–2715 (1997).
Tran, P.H., Korszun, Z.R., Cerritelli, S., Springhorn, S.S. & Lacks, S.A. Crystal structure of the DpnM DNA adenine methyltransferase from the DpnII restriction system of streptococcus pneumoniae bound to S-adenosylmethionine. Structure 6, 1563–1575 (1998).
Scavetta, R.D. et al. Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases. Nucleic Acids Res. 28, 3950–3961 (2000).
Schubert, H.L., Blumenthal, R.M. & Cheng, X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 28, 329–335 (2003).
Beck, C. & Jeltsch, A. Probing the DNA interface of the EcoRV DNA-(adenine-N6)-methyltransferase by site-directed mutagenesis, fluorescence spectroscopy, and UV cross-linking. Biochemistry 41, 14103–14110 (2002).
Miner, Z., Schlagman, S.L. & Hattman, S. Single amino acid changes that alter the DNA sequence specificity of the DNA-[N6-adenine] methyltransferase (Dam) of bacteriophage T4. Nucleic Acids Res. 17, 8149–8157 (1989).
Hattman, S. DNA methylation of T-even bacteriophages and of their nonglucosylated mutants: its role in P1-directed restriction. Virology 42, 359–367 (1970).
Minko, I., Hattman, S., Lloyd, R.S. & Kossykh, V. Methylation by a mutant T2 DNA [N(6)-adenine] methyltransferase expands the usage of RecA-assisted endonuclease (RARE) cleavage. Nucleic Acids Res. 29, 1484–1490 (2001).
Kossykh, V.G., Schlagman, S.L. & Hattman, S. Conserved sequence motif DPPY in region IV of the phage T4 Dam DNA-[N6-adenine]-methyltransferase is important for S-adenosyl-L-methionine binding. Nucleic Acids Res. 21, 4659–4662 (1993).
Schluckebier, G., O'Gara, M., Saenger, W. & Cheng, X. Universal catalytic domain structure of AdoMet-dependent methyltransferases. J. Mol. Biol. 247, 16–20 (1995).
Tuzikov, F.V., Tuzikova, N.A., Naumochkin, A.N., Zinov'ev, V.V. & Malygin, E.G. Fluorescence quenching study of the equilibrium binding of phage T4 DamDNA-[N6-adenine]-methyltransferase with substrates and ligands. Mol. Biol. (Mosk) 31, 73–76 (1997).
Roth, M. & Jeltsch, A. Changing the target base specificity of the EcoRV DNA methyltransferase by rational de novo protein-design. Nucleic Acids Res. 29, 3137–3144 (2001).
Malygin, E.G. et al. A dual role for substrate S-adenosyl-L-methionine in the methylation reaction with bacteriophage T4 Dam DNA-[N6-adenine]-methyltransferase. Nucleic Acids Res. 29, 2361–2369 (2001).
Murphy, F.V.T. & Churchill, M.E. Nonsequence-specific DNA recognition: a structural perspective. Structure 8, R83–R89 (2000).
Lee, B. & Richards, F.M. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 55, 379–400 (1971).
Kleywegt, G.J. & Jones, T.A. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D 50, 178–185 (1994).
Urig, S. et al. The Escherichia coli dam DNA methyltransferase modifies DNA in a highly processive reaction. J. Mol. Biol. 319, 1085–1096 (2002).
Zinoviev, V.V., Evdokimov, A.A., Malygin, E.G., Schlagman, S.L. & Hattman, S. Bacteriophage T4 Dam DNA-[N6-adenine]-methyltransferase: processivity and orientation to the methylation target. J. Biol. Chem. 278, 7829–7833 (2002).
Evdokimov, A.A., Zinoviev, V.V., Malygin, E.G., Schlagman, S.L. & Hattman, S. Bacteriophage T4 Dam DNA-[N6-adenine]methyltransferase. Kinetic evidence for a catalytically essential conformational change in the ternary complex. J. Biol. Chem. 277, 279–286 (2002).
Malygin, E.G., Lindstrom, W.M., Jr., Schlagman, S.L., Hattman, S. & Reich, N.O. Pre-steady state kinetics of bacteriophage T4 Dam DNA-[N(6)-adenine] methyltransferase: interaction with native (GATC) or modified sites. Nucleic Acids Res. 28, 4207–4211 (2000).
Breyer, W.A. & Matthews, B.W. A structural basis for processivity. Protein Sci. 10, 1699–1711 (2001).
Kossykh, V.G., Schlagman, S.L. & Hattman, S. Phage T4 DNA [N6-adenine]methyltransferase. Overexpression, purification, and characterization. J. Biol. Chem. 270, 14389–14393 (1995).
Terwilliger, T.C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999).
Terwilliger, T.C. Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 (2000).
Furey, W. & Swaminathan, S. PHASES-95: A program package for processing and analyzing diffraction data from macromolecules. Methods Enzymol. 277, 590–620 (1997).
Abrahams, J.P. et al. The structure of bovine F1-ATPase complexed with the peptide antibiotic efrapeptin. Proc. Natl. Acad. Sci. USA 93, 9420–9424 (1996).
Cowtan, K.D. & Zhang, K.Y. Density modification for macromolecular phase improvement. Prog. Biophys. Mol. Biol. 72, 245–270 (1999).
Brunger, A.T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Navaza, J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D 57, 1367–1372 (2001).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Carson, M. Ribbons. Methods Enzymol. 227, 493–505 (1997).
Nicholls, A., Sharp, K.A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991).
Miner, Z. & Hattman, S. Molecular cloning, sequencing, and mapping of the bacteriophage T2 dam gene. J. Bacteriol. 170, 5177–5184 (1988).
Kossykh, V.G., Schlagman, S.L. & Hattman, S. Comparative studies of the phage T2 and T4 DNA (N6-adenine)methyltransferases: amino acid changes that affect catalytic activity. J. Bacteriol. 179, 3239–3243 (1997).
Acknowledgements
We thank beamline staff for help with X-ray data collection at beamlines X12C, X25 and X26C in the facilities of the National Synchrotron Light Source, Brookhaven National Laboratory, R.M. Blumenthal (Medical College of Ohio) for comments and M. Churchill (University of Colorado) for discussion. These studies were supported in part by US Public Health Services grants to S.H and X.C. and the Georgia Research Alliance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Yang, Z., Horton, J., Zhou, L. et al. Structure of the bacteriophage T4 DNA adenine methyltransferase. Nat Struct Mol Biol 10, 849–855 (2003). https://doi.org/10.1038/nsb973
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb973
This article is cited by
-
Two novel phages, Klebsiella phage GADU21 and Escherichia phage GADU22, from the urine samples of patients with urinary tract infection
Virus Genes (2024)
-
Clostridioides difficile specific DNA adenine methyltransferase CamA squeezes and flips adenine out of DNA helix
Nature Communications (2021)
-
The cell cycle-regulated DNA adenine methyltransferase CcrM opens a bubble at its DNA recognition site
Nature Communications (2019)
-
Methyltransferases acquired by lactococcal 936-type phage provide protection against restriction endonuclease activity
BMC Genomics (2014)
-
The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature (2008)