Abstract
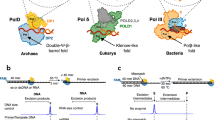
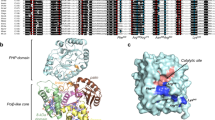
Deamination of cytosine to uracil in a G-C base pair is a major promutagenic event, generating G-C→A-T mutations if not repaired before DNA replication. Archaeal family B DNA polymerases are uniquely able to recognize unrepaired uracil in a template strand and stall polymerization upstream of the lesion, thereby preventing the irreversible fixation of an A-T mutation. We have now identified a 'pocket' in the N-terminal domains of archaeal DNA polymerases that is positioned to interact with the template strand and provide this ability. The structure of this pocket provides interacting groups that discriminate uracil from the four normal DNA bases (including thymine). These groups are conserved in archaeal polymerases but absent from homologous viral polymerases that are unable to recognize uracil. Using site-directed mutagenesis, we have confirmed the biological role of this pocket and have engineered specific mutations in the Pfu polymerase that confer the ability to read through template-strand uracils and carry out PCR with dUTP in place of dTTP.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lindahl, T. Nature 362, 709–715 (1993).
Krokan, H.E., Standal, R. & Slupphaug, G. Biochem. J. 325, 1–16 (1997).
Parikh, S.S., Mol, C.D., Hosfield, D.J. & Tainer, J.A. Curr. Opin. Struct. Biol. 9, 37–47 (1999).
Pearl, L.H. Mutat. Res. 460, 165–181 (2000).
Lindahl, T. Proc. Natl. Acad. Sci. USA 71, 3649–3653 (1974).
Lindahl, T. & Nyberg, B. Biochemistry 13, 3405–3410 (1974).
Grogan, D.W. Trends Microbiol. 8, 180–185 (2000).
Horst, J.P. & Fritz, H.J. EMBO J. 15, 5459–5469 (1996).
Yang, H. et al. J. Bacteriol. 182, 1272–1279 (2000).
Sandigursky, M. & Franklin, W.A. J. Biol. Chem. 275, 19146–19149 (2000).
Sartori, A.A., Schär, P., Fitz-Gibbon, S., Miller, J.E. & Jiriciny, J. J. Biol. Chem. 276, 29979–29986 (2001).
Sandigursky, M. & Franklin, W.A. Curr. Biol. 9, 531–534 (1999).
Sandigursky, M., Faje, A. & Franklin, W.A. Mutat. Res. 485, 187–195 (2001).
Lasken, R.S., Schuster, D.M. & Rashtchian, A. J. Biol. Chem. 271, 17692–17696 (1996).
Greagg, M.A. et al. Proc. Natl. Acad. Sci. USA 96, 9045–9050 (1999).
Shinohara, A. & Ogawa, T. Trends Biochem. Sci. 20, 387–391 (1995).
Fuchs, R.P.P. & Baynton, K. Trends Biochem. Sci. 25, 74–79 (2000).
Wang, J. et al. Cell 89, 1087–1099 (1997).
Franklin, M.C., Wang, J. & Steitz, T.A. Cell 105, 657–667 (2001).
Hopfner, K.P. et al. Proc. Natl. Acad. Sci. USA 96, 3600–3605 (1999).
Zhao, Y. et al. Structure 7, 1189–1199 (1999).
Rodriguez, A.C., Park, H.-W., Mao, C. & Beese, L. J. Mol. Biol. 299, 447–462 (2000).
Hashimoto, H. et al. J. Mol. Biol. 306, 469–477 (2001).
Wang, J., Yu, P., Lin, T.C., Konigsberg, W.H. & Steitz, T.A. Biochemistry 35, 8110–8119 (1996).
Reid, S.L., Parry, D., Liu, H.-H. & Connolly, B.A. Biochemistry 40, 2484–2484 (2001).
Hogrefe, H.H., Hansen, C.J., Scott, B.R. & Nielson, K.B. Proc. Natl. Acad. Sci. USA 99, 596–602 (2002).
Larson, G., Svenson, L.A. & Nyman, P.O. Nature Struct. Biol. 3, 532–536 (1996).
Pearl, L.H. & Savva, R. Nature Struct. Biol. 3, 485–487 (1996).
Ronen, A. & Glickman, B.W. Environ. Mol. Mutagen. 37, 241–283 (2001).
Wood, R.D., Mitchell, M., Sgouros, J. & Lindahl, T. Science, 291, 1284–1289 (2001).
Goodman, M.F. Trends Biochem. Sci. 25, 189–195 (2000).
Evans S.J. et al. Nucleic Acids. Res. 28, 1059–1066 (2000).
Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K. & Pease, L.R. Gene 77, 51–59 (1989).
Richardson, C.C. In Procedures in Nucleic Acids Research (eds Cantoni, G.L. & Davies, D.R.) 263–276 (Harper and Row, New York; 1966).
Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F. & Higgins, D.G. Nucleic Acids Res. 24, 4876–4882 (1997).
Acknowledgements
This work was supported by the UK Biotechnology and Biological Science Research Council, Cancer Research UK and the UK Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Some of the mutants described in this paper, which may be useful in PCR have been patented.
Rights and permissions
About this article
Cite this article
Fogg, M., Pearl, L. & Connolly, B. Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat Struct Mol Biol 9, 922–927 (2002). https://doi.org/10.1038/nsb867
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb867
This article is cited by
-
Animal species identification utilising DNAs extracted from traditionally manufactured gelatin (Wanikawa)
Heritage Science (2022)
-
Isolation and engineering of a Listeria grayi bacteriophage
Scientific Reports (2021)
-
Identification of Thermus aquaticus DNA polymerase variants with increased mismatch discrimination and reverse transcriptase activity from a smart enzyme mutant library
Scientific Reports (2019)
-
TT(N)mGCCTC inhibits archaeal family B DNA polymerases
Scientific Reports (2018)
-
PCNA is involved in the EndoQ-mediated DNA repair process in Thermococcales
Scientific Reports (2016)