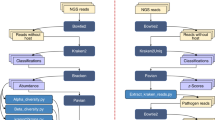
Abstract
The development of affordable, high-throughput sequencing technology has led to a flood of publicly available bacterial genome-sequence data. The availability of multiple genome sequences presents both an opportunity and a challenge for microbiologists, and new computational approaches are needed to extract the knowledge that is required to address specific biological problems and to analyse genomic data. The field of e-Science is maturing, and Grid-based technologies can help address this challenge.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Luciano, J. S. & Stevens, R. D. e-Science and biological pathway semantics. BMC Bioinformatics 8, S3 (2007).
de Roure, D., Goble, C. & Stevens, R. in Proc. 2007 IEEE Conf. eScience Grid Comput. 603–610 (2007).
Foster, I., Kesselman, C. & Tuecke, S. The anatomy of the Grid: enabling scalable virtual organizations. Int. J. High Perform. Comput. Appl. 15, 200–222 (2001).
Foster, I. & Kesselman, C. Globus: a metacomputing infrastructure toolkit. Int. J. High Perform. Comput. Appl. 11, 115–128 (1997).
Thain, D., Tannenbaum, T. & Livny, M. in Grid Computing (eds Berman, F., Fox, G. & Hey, T.) 299–335 (2003).
Stajich, J. E. et al. The Bioperl toolkit: perl modules for the life sciences. Genome Res. 12, 1611–1618 (2002).
Chapman, B. & Chang, J. Biopython: Python tools for computational biology. ACM SIGBIO Newsl. 20, 15–19 (2000).
Pocock, M., Down, T. & Hubbard, T. BioJava: open source components for bioinformatics. ACM SIGBIO Newsl. 20, 10–12 (2000).
Oinn, T. et al. Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics 20, 3045–3054 (2004).
Stevens, R. D., Robinson, A. J. & Goble, C. A. myGrid: personalised bioinformatics on the information grid. Bioinformatics 19, I302–I304 (2003).
Curbera, F. et al. Unraveling the web services web: an introduction to SOAP, WSDL, and UDDI. IEEE Internet Comput. 6, 86–93 (2002).
Khare, R. & Taylor, R. N. in Proc. 26th Int. Conf. Software Eng. (ed. Taylor, R. N.) 428–437 (2004).
Wilkinson, M. D. & Links, M. BioMOBY: an open source biological web services proposal. Brief. Bioinformatics 3, 331–341 (2002).
Foster, I., Kesselman, C., Nick, J. M. & Tuecke, S. Grid services for distributed system integration. Computer 35, 37–46 (2002).
Hull, D. et al. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 34, W729–W732 (2006).
Pillai, S. et al. SOAP-based services provided by the European Bioinformatics Institute. Nucleic Acids Res. 33, W25–W28 (2005).
Senger, M., Rice, P. & Oinn, T. in UK e-Science All Hands Meet. 2003 (ed. Cox, S. J.) 509–513 (2003).
Majithia, S., Shields, M., Taylor, I. & Wang, I. in Proc. IEEE Intern. Conf. Web Services (ed. Shields, M.) 514–521 (2004).
Castro, A. G., Thoraval, S., Garcia, L. J. & Ragan, M. A. Workflows in bioinformatics: meta-analysis and prototype implementation of a workflow generator. BMC Bioinform. 6, 87 (2005).
Ludäscher, B. et al. Scientific workflow management and the kepler system. Concurr. Comput. Pract. Exper. 18, 1039–1065 (2006).
Stevens, R. et al. myGrid and the drug discovery process. Drug Discov. Today 2, 140–148 (2004).
Fisher, P. et al. A systematic strategy for large-scale analysis of genotype phenotype correlations: identification of candidate genes involved in African trypanosomiasis. Nucleic Acids Res. 35, 5625–5633 (2007).
Agostini, F. P., Soares-Pinto, D. O., Moret, M. A., Osthoff, C. & Pascutti, P. G. Generalized simulated annealing applied to protein folding studies. J. Comput. Chem. 11, 1142–1152 (2006).
Craddock, T., Lord, P., Harwood, C. R. & Wipat, A. in Proc. 5th UK e-Science All Hands Meet. 788–795 (2006).
Harwood, C. R. & Cranenburgh, R. Bacillus protein secretion: an unfolding story. Trends Microbiol. 16, 73–79 (2008).
Juncker, A. S. et al. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12, 1652–1662 (2003).
Bendtsen, J. D., Nielsen, H., von Heijne, G. & Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 (2004).
Sonnhammer, E. L. L., von Heijne, G. & Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182 (1998).
Jones, D. T., Taylor, W. R. & Thornton, J. M. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33, 3038–3049 (1994).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Enright, A. J., Kunin, V. & Ouzounis, C. A. Protein families and TRIBES in genome sequence space. Nucleic Acids Res. 31, 4632–4638 (2003).
Sun, Y. et al. Exploring microbial genome sequences to identify protein families on the Grid. IEEE Trans. Inf. Technol. Biomed., 11 (2007).
Sun, Y. et al. in 2005 IEEE International Symposium on Cluster Computing and the Grid 977–984 (2005).
Hedeler, C. et al. e-Fungi: a data resource for comparative analysis of fungal genomes. BMC Genomics 8, 426 (2007).
Chaudhuri, R. R. et al. xBASE2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res. 36, D543–D546 (2008).
Sulakhe, D. et al. Gnare: automated system for high-throughput genome analysis with Grid computational backend. J. Clin. Monit. Comput. 19, 361–369 (2005).
Sulakhe, D., Rodriguez, A., Wilde, M., Foster, I. A. & Maltsev, N. A. Interoperability of GADU in using heterogeneous grid resources for bioinformatics applications. IEEE Trans. Inf. Technol. Biomed. 12, 241–246 (2008).
Seshadri, R., Kravitz, S. A., Smarr, L., Gilna, P. & Frazier, M. CAMERA: a community resource for metagenomics. PLoS Biol. 5, e75 (2007).
Maltsev, N. A. et al. PUMA2-grid-based high-throughput analysis of genomes and metabolic pathways. Nucleic Acids Res. 34, D369–D372 (2006).
Schulze-Kremer, S. Ontologies for molecular biology. in Proc. 3rd Pacific Symp. Biocomput., 693–704 (1998).
Kohler, J. et al. Graph-based analysis and visualization of experimental results with ONDEX. Bioinformatics 22, 1383–1390 (2006).
Papanikou, E., Karamanou, S. & Economou, A. Bacterial protein secretion through the translocase nanomachine. Nature Rev. Microbiol. 5, 839–851 (2007).
Berks, B. C., Palmer, T. & Sargent, F. Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr. Opin. Microbiol. 8, 174–181 (2005).
Acknowledgements
The authors acknowledge funding from the UK Engineering and Physical Sciences Research Council and Non-linear Dynamics for a CASE (collaborative awards in science and engineering) studentship to T.C., from Research Councils UK for a fellowship to J.H. and from the European Union (Bacell Health; grant number LSH-2002-1.1.0-1).
Author information
Authors and Affiliations
Corresponding author
Related links
Related links
DATABASES
Entrez Genome Project
FURTHER INFORMATION
EMBL–EBI (Genome Pages — Bacteria)
Rights and permissions
About this article
Cite this article
Craddock, T., Harwood, C., Hallinan, J. et al. e-Science: relieving bottlenecks in large-scale genome analyses. Nat Rev Microbiol 6, 948–954 (2008). https://doi.org/10.1038/nrmicro2031
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2031
This article is cited by
-
Reducing data transfer in big-data workflows: the computation-flow delegated approach
Journal of Data, Information and Management (2019)
-
e!DAL - a framework to store, share and publish research data
BMC Bioinformatics (2014)
-
Initial steps towards a production platform for DNA sequence analysis on the grid
BMC Bioinformatics (2010)
-
Applications of thiol-disulfide oxidoreductases for optimized in vivo production of functionally active proteins in Bacillus
Applied Microbiology and Biotechnology (2009)