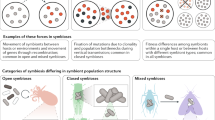
Key Points
-
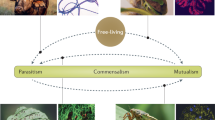
Microbial symbiosis is a central factor in the evolution of all animal species and in the survival of all individuals. Advances in molecular genetics are allowing the development of invertebrate–bacteria associations as model systems for understanding how the host and microorganism communicate.
-
Five natural systems have been particularly well described, including three that are binary (the host and one bacterial species) and two that consist of simple bacterial consortia of fewer than a dozen species.
-
The development of these experimental symbiosis models has allowed us to ask distinct sets of questions about host–microorganism communication and has provided a breadth of opportunities that would not be available from any one system.
-
In the past 20 years, advances in our understanding of host–microorganism communication have centred on several common themes: surface structures and specificity; bacterial behaviour and gene regulation; adaptation to host defences; induction of host development; and nutritional and metabolic accommodation. Although parallels can be drawn between the systems, there are also intriguing differences that reflect the special biology of each host–microorganism association.
-
A number of newly recognized symbioses between bacteria and invertebrate hosts are emerging as model systems in which molecular genetic approaches are only now being applied.
Abstract
The recent development and application of molecular genetics to the symbionts of invertebrate animal species have advanced our knowledge of the biochemical communication that occurs between the host and its bacterial symbionts. In particular, the ability to manipulate these associations experimentally by introducing genetic variants of the symbionts into naive hosts has allowed the discovery of novel colonization mechanisms and factors. In addition, the role of the symbionts in inducing normal host development has been revealed, and its molecular basis described. In this Review, I discuss many of these developments, focusing on what has been discovered in five well-understood model systems.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Backhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A. & Gordon, J. I. Host–bacterial mutualism in the human intestine. Science 307, 1915–1920 (2005).
Haygood, M. G. Light organ symbioses in fishes. Crit. Rev. Microbiol. 19, 191–216 (1993).
Parniske, M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nature Rev. Microbiol. 6, 763–775 (2008)
Werren, J. H., Baldo, L. & Clark, M. E. Wolbachia: master manipulators of invertebrate biology. Nature Rev. Microbiol. 6, 741–751 (2008).
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R. & Gordon, J. I. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Rev. Microbiol. 6, 776–788 (2008).
Dethlefsen, L., McFall-Ngai, M. & Relman, D. A. An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature 449, 811–818 (2007).
Ley, R. E., Peterson, D. A. & Gordon, J. I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848 (2006).
Samuel, B. S. & Gordon, J. I. A humanized gnotobiotic mouse model of host–archaeal–bacterial mutualism. Proc. Natl Acad. Sci. USA 103, 10011–10016 (2006).
Nicholson, J. K., Holmes, E. & Wilson, I. D. Gut microorganisms, mammalian metabolism and personal heath care. Nature Rev. Microbiol. 3, 431–438 (2005).
Peterson, D. A., McNulty, N. P., Guruge, J. L. & Gordon, J. I. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2, 328–339 (2007).
Turnbaugh, P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006).
Vaishnava, S., Behrendt, C. L. & Hooper, L. V. Innate immune responses to commensal bacteria in the gut epithelium. J. Pediatr. Gastroenterol. Nutr. 46 (Suppl. 1), E10–E11 (2008).
Graf, J., Kikuchi, Y. & Rio, R. V. Leeches and their microbiota: naturally simple symbiosis models. Trends Microbiol. 14, 365–371 (2006).
Cox, C. R. & Gilmore, M. S. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect. Immun. 75, 1565–1576 (2007).
McFall-Ngai, M. J. & Gordon, J. I. in Evolution of Microbial Virulence (eds Seifert, H. & DiRita, V. J.) 147–166 (ASM, Washington DC, 2006).
Goodrich-Blair, H. & Clarke, D. J. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination. Mol. Microbiol. 64, 260–268 (2007).
McFall-Ngai, M. Adaptive immunity: care for the community. Nature 445, 153 (2007).
Moran, N. A. Bacterial menageries inside insects. Proc. Natl Acad. Sci. USA 98, 1338–1340 (2001).
Nealson, K. H. & Hastings, J. W. in The Prokaryotes, a Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications 2nd edn (eds Balows, A., Truper, H. G., Dworkin, M., Harder, W. & Schleifer, K. H.) 1332–1345 (Springer, Berlin, 1991).
Fidopiastis, P. M., von Boletzky, S. & Ruby, E. G. A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J. Bacteriol. 180, 59–64 (1998).
Nishiguchi, M. K. & Nair, V. S. Evolution of symbiosis in the Vibrionaceae: a combined approach using molecules and physiology. Int. J. Syst. Evol. Microbiol. 53, 2019–2026 (2003).
Jones, B. W. & Nishiguchi, M. K. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca:Cephalopoda). Mar. Biol. 144, 1151–1155 (2004).
Wei, S. L. & Young, R. E. Development of symbiotic bacterial luminescence in a nearshore cephalopod, Euprymna scolopes. Mar. Biol. 103, 541–546 (1989).
Ruby, E. G. & Lee, K. H. The Vibrio fischeri–Euprymna scolopes light organ association: current ecological paradigms. Appl. Environ. Microbiol. 64, 805–812 (1998).
Claes, M. F. & Dunlap, P. V. Aposymbiotic culture of the sepiolid squid Euprymna scolopes: role of the symbiotic bacterium Vibrio fischeri in host animal growth, development, and light organ morphogenesis. J. Exp. Zool. 286, 280–296 (2000).
Dunn, A. K., Millikan, D. S., Adin, D. M., Bose, J. L. & Stabb, E. V. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 72, 802–810 (2006).
Stabb, E. V. & Ruby, E. G. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358, 413–426 (2002).
Mandel, M. J., Stabb, E. V. & Ruby, E. G. Comparative genomics-based investigation of resequencing targets in Vibrio fischeri: focus on point miscalls and artefactual expansions. BMC Genomics 9, 138 (2008). Introduced novel technological approaches to apply a comparative genomics approach to two strains of a beneficial bacterial symbiont.
Ruby, E. G. et al. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl Acad. Sci. USA 102, 3004–3009 (2005). Sequencing of the V. fischeri genome, which allowed molecular genetics to be applied to the squid– Vibrio system and opened up new approaches of genetic analysis.
Antunes, L. C. et al. Transcriptome analysis of the Vibrio fischeri LuxR–LuxI regulon. J. Bacteriol. 189, 8387–8391 (2007).
Lupp, C. & Ruby, E. G. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol. 187, 3620–3629 (2005).
Chun, C. K. et al. An annotated cDNA library of juvenile Euprymna scolopes with and without colonization by the symbiont Vibrio fischeri. BMC Genomics 7, 154 (2006).
Chun, C. et al. Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid–vibrio association. Proc. Natl Acad. Sci. USA 5 Aug 2008 (doi:10.1073/pnas.0802369105). First large-scale transcriptional analysis of the host response to colonization by bacteria that possess mutations in their symbiosis genes.
Herbert, E. E. & Goodrich-Blair, H. Friend and foe: the two faces of Xenorhabdus nematophila. Nature Rev. Microbiol. 5, 634–646 (2007).
Hallem, E. A., Rengarajan, M., Ciche, T. A. & Sternberg, P. W. Nematodes, bacteria, and flies: a tripartite model for nematode parasitism. Curr. Biol. 17, 898–904 (2007).
Ciche, T. A., Kim, K. S., Kaufmann-Daszczuk, B., Nguyen, K. C. & Hall, D. H. Cell invasion and matricide during Photorhabdus luminescens transmission by Heterorhabditis bacteriophora nematodes. Appl. Environ. Microbiol. 74, 2275–2287 (2008). The first study to focus on development of the host during an association with a nematode.
Cowles, C. E. & Goodrich-Blair, H. The Xenorhabdus nematophila nilABC genes confer the ability of Xenorhabdus spp. to colonize Steinernema carpocapsae nematodes. J. Bacteriol. 190, 4121–4128 (2008).
Goodrich-Blair, H. They've got a ticket to ride: Xenorhabdus nematophila–Steinernema carpocapsae symbiosis. Curr. Opin. Microbiol. 10, 225–230 (2007).
Joyce, S. A., Watson, R. J. & Clarke, D. J. The regulation of pathogenicity and mutualism in Photorhabdus. Curr. Opin. Microbiol. 9, 127–132 (2006).
Dale, C. & Moran, N. A. Molecular interactions between bacterial symbionts and their hosts. Cell 126, 453–465 (2006).
Moran, N. A. Symbiosis. Curr. Biol. 16, R866–R871 (2006).
Toh, H. et al. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 16, 149–156 (2006).
Weiss, B. L. et al. Interspecific transfer of bacterial endosymbionts between tsetse fly species: infection establishment and effect on host fitness. Appl. Environ. Microbiol. 72, 7013–7021 (2006).
Rio, R. V., Lefevre, C., Heddi, A. & Aksoy, S. Comparative genomics of insect-symbiotic bacteria: influence of host environment on microbial genome composition. Appl. Environ. Microbiol. 69, 6825–6832 (2003).
Warnecke, F. et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450, 560–565 (2007).
Broderick, N. A., Raffa, K. F., Goodman, R. M. & Handelsman, J. Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl. Environ. Microbiol. 70, 293–300 (2004).
Indergand, S. & Graf, J. Ingested blood contributes to the specificity of the symbiosis of Aeromonas veronii biovar sobria and Hirudo medicinalis, the medicinal leech. Appl. Environ. Microbiol. 66, 4735–4741 (2000).
Silver, A. C. et al. Interaction between innate immune cells and a bacterial type III secretion system in mutualistic and pathogenic associations. Proc. Natl Acad. Sci. USA 104, 9481–9486 (2007). Discovery of a T3SS in a beneficial bacteria–animal symbiosis, and described one of its functions.
Silver, A. C., Rabinowitz, N. M., Kuffer, S. & Graf, J. Identification of Aeromonas veronii genes required for colonization of the medicinal leech, Hirudo verbana. J. Bacteriol. 189, 6763–6772 (2007).
Kikuchi, Y. & Graf, J. Spatial and temporal population dynamics of a naturally occurring two-species microbial community inside the digestive tract of the medicinal leech. Appl. Environ. Microbiol. 73, 1984–1991 (2007).
Broderick, N. A., Raffa, K. F. & Handelsman, J. Midgut bacteria required for Bacillus thuringiensis insecticidal activity. Proc. Natl Acad. Sci. USA 103, 15196–15199 (2006).
Falkow, S. Molecular Koch's postulates applied to bacterial pathogenicity — a personal recollection 15 years later. Nature Rev. Microbiol. 2, 67–72 (2004).
Heungens, K., Cowles, C. E. & Goodrich-Blair, H. Identification of Xenorhabdus nematophila genes required for mutualistic colonization of Steinernema carpocapsae nematodes. Mol. Microbiol. 45, 1337–1353 (2002). One of the first applications of an advanced genetic screen for bacterial colonization factors in an animal symbiont.
Nyholm, S. V., Stabb, E. V., Ruby, E. G. & McFall-Ngai, M. J. Establishment of an animal–bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl Acad. Sci. USA 97, 10231–10235 (2000).
Hussa, E. A., O'Shea, T. M., Darnell, C. L., Ruby, E. G. & Visick, K. L. Two-component response regulators of Vibrio fischeri: identification, mutagenesis, and characterization. J. Bacteriol. 189, 5825–5838 (2007).
Yip, E. S., Geszvain, K., Deloney-Marino, C. R. & Visick, K. L. The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol. Microbiol. 52, 1586–1600 (2006). Provided a breakthrough in our understanding of the regulation of genes that are involved in symbiosis initiation in the squid– Vibrio association.
Darnell, C. L., Hussa, E. A. & Visick, K. L. The putative hybrid sensor kinase SypF coordinates biofilm formation in Vibrio fischeri by acting upstream of two response regulators, SypG and VpsR. J. Bacteriol. 190, 4941–4950 (2008).
Geszvain, K. & Visick, K. L. Roles of bacterial regulators in the symbiosis between Vibrio fischeri and Euprymna scolopes. Prog. Mol. Subcell. Biol. 41, 277–290 (2006).
DeLoney, C. R., Bartley, T. M. & Visick, K. L. Role for phosphoglucomutase in Vibrio fischeri–Euprymna scolopes symbiosis. J. Bacteriol. 184, 5121–5129 (2002).
Adin, D. M. et al. Characterization of htrB and msbB mutants of the light organ symbiont Vibrio fischeri. Appl. Environ. Microbiol. 74, 633–644 (2007).
Gunn, J. S., Ryan, S. S., Van Velkinburgh, J. C., Ernst, R. K. & Miller, S. I. Genetic and functional analysis of a PmrA–PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 68, 6139–6146 (2000).
Bennett, H. P. & Clarke, D. J. The pbgPE operon in Photorhabdus luminescens is required for pathogenicity and symbiosis. J. Bacteriol. 187, 77–84 (2005).
Gunn, J. S. et al. PmrA–PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27, 1171–1182 (1998).
Braschler, T. R., Merino, S., Tomas, J. M. & Graf, J. Complement resistance is essential for colonization of the digestive tract of Hirudo medicinalis by Aeromonas strains. Appl. Environ. Microbiol. 69, 4268–4271 (2003).
Aeckersberg, F., Lupp, C., Feliciano, B. & Ruby, E. G. Vibrio fischeri outer membrane protein OmpU plays a role in normal symbiotic colonization. J. Bacteriol. 183, 6590–6597 (2001).
Stabb, E. V. & Ruby, E. G. Contribution of pilA to competitive colonization of the squid Euprymna scolopes by Vibrio fischeri. Appl. Environ. Microbiol. 69, 820–826 (2003).
Lee, K.-H. & Ruby, E. G. Detection of the light organ symbiont, Vibrio fischeri, in Hawaiian seawater by using lux gene probes. Appl. Environ. Microbiol. 58, 942–947 (1992).
Lee, K.-H. & Ruby, E. G. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl. Environ. Microbiol. 60, 1565–1571 (1994).
He, H., Snyder, H. A. & Forst, S. Unique organization and regulation of the mrx fimbrial operon in Xenorhabdus nematophila. Microbiology 150, 1439–1446 (2004).
Fuqua, C. & Greenberg, E. P. Listening in on bacteria: acyl-homoserine lactone signalling. Nature Rev. Mol. Cell Biol. 3, 685–695 (2002).
Alegado, R. A., Campbell, M. C., Chen, W. C., Slutz, S. S. & Tan, M. W. Characterization of mediators of microbial virulence and innate immunity using the Caenorhabditis elegans host–pathogen model. Cell Microbiol. 5, 435–444 (2003).
Lupp, C. & Ruby, E. G. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J. Bacteriol. 186, 3873–3881 (2004).
Studer, S. V., Mandel, M. J. & Ruby, E. G. AinS quorum sensing regulates the Vibrio fischeri acetate switch. J. Bacteriol. 190, 5915–5923 (2008).
Wolfe, A. J. The acetate switch. Microbiol. Mol. Biol. Rev. 69, 12–50 (2005).
Callahan, S. M. & Dunlap, P. V. LuxR- and acyl-homoserine-lactone-controlled non-lux genes define a quorum-sensing regulon in Vibrio fischeri. J. Bacteriol. 182, 2811–2822 (2000).
Visick, K. L., Foster, J., Doino, J., McFall-Ngai, M. & Ruby, E. G. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182, 4578–4586 (2000). A key paper that links the production of luminescence to both the induction of normal host development and the capacity for persistent colonization.
Boettcher, K. J. & Ruby, E. G. Detection and quantification of Vibrio fischeri autoinducer from symbiotic squid light organs. J. Bacteriol. 177, 1053–1058 (1995).
Sanchez-Contreras, M., Bauer, W. D., Gao, M., Robinson, J. B. & Allan Downie, J. Quorum-sensing regulation in rhizobia and its role in symbiotic interactions with legumes. Philos. Trans. R. Soc. Lond. B 362, 1149–1163 (2007).
You, J., Liang, S., Cao, L., Liu, X. & Han, R. Nutritive significance of crystalline inclusion proteins of Photorhabdus luminescens in Steinernema nematodes. FEMS Microbiol. Ecol. 55, 178–185 (2006).
Joyce, S. A. & Clarke, D. J. A hexA homologue from Photorhabdus regulates pathogenicity, symbiosis and phenotypic variation. Mol. Microbiol. 47, 1445–1457 (2003).
Wolfe, A. J., Millikan, D. S., Campbell, J. M. & Visick, K. L. Vibrio fischeri σ54 controls motility, biofilm formation, luminescence, and colonization. Appl. Environ. Microbiol. 70, 2520–2524 (2004).
DeLoney-Marino, C. R., Wolfe, A. J. & Visick, K. L. Chemoattraction of Vibrio fischeri to serine, nucleosides, and N-acetylneuraminic acid, a component of squid light-organ mucus. Appl. Environ. Microbiol. 69, 7527–7530 (2003).
Graf, J., Dunlap, P. V. & Ruby, E. G. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 176, 6986–6991 (1994).
Graf, J. & Ruby, E. G. Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence, as well as nitrogen utilization. Mol. Microbiol. 37, 168–179 (2000).
Bose, J. L. et al. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol. Microbiol. 65, 538–553 (2007).
Dale, C., Young, S. A., Haydon, D. T. & Welburn, S. C. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc. Natl Acad. Sci. USA 98, 1883–1888 (2001).
Visick, K. L. & Ruby, E. G. TnluxAB insertion mutants of Vibrio fischeri with symbiosis-regulated phenotypes. Nova Acta Leopoldina 333, 93–100 (2003).
Martens, E. C., Russell, F. M. & Goodrich-Blair, H. Analysis of Xenorhabdus nematophila metabolic mutants yields insight into stages of Steinernema carpocapsae nematode intestinal colonization. Mol. Microbiol. 58, 28–45 (2005).
Davidson, S. K., Koropatnick, T. A., Kossmehl, R., Sycuro, L. & McFall-Ngai, M. J. NO means 'yes' in the squid–vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell. Microbiol. 6, 1139–1151 (2004).
Visick, K. L. & Ruby, E. G. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and approach to stationary phase. J. Bacteriol. 180, 2087–2092 (1998).
Krin, E. et al. Pleiotropic role of quorum-sensing autoinducer 2 in Photorhabdus luminescens. Appl. Environ. Microbiol. 72, 6439–6451 (2006).
Rio, R. V., Anderegg, M. & Graf, J. Characterization of a catalase gene from Aeromonas veronii, the digestive-tract symbiont of the medicinal leech. Microbiology 153, 1897–1906 (2007).
Dale, C., Jones, T. & Pontes, M. Degenerative evolution and functional diversification of type-III secretion systems in the insect endosymbiont Sodalis glossinidius. Mol. Biol. Evol. 22, 758–766 (2005).
Brugirard-Ricaud, K. et al. Site-specific antiphagocytic function of the Photorhabdus luminescens type III secretion system during insect colonization. Cell. Microbiol. 7, 363–371 (2005).
Nyholm, S. V. & McFall-Ngai, M. J. The winnowing: establishing the squid-Vibrio symbiosis. Nature Rev. Microbiol. 2, 632–642 (2004).
Stappenbeck, T. S., Hooper, L. V. & Gordon, J. I. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl Acad. Sci. USA 99, 15451–15455 (2002).
Ruby, E. G. & McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends Microbiol. 7, 414–420 (1999).
Koropatnick, T. A. et al. Microbial factor-mediated development in a host–bacterial mutualism. Science 306, 1186–1188 (2004). Developed a new paradigm by showing that a bacterial toxin serves as a required developmental signal compound in a beneficial host–microorganism association.
Whistler, C. A. & Ruby, E. G. GacA regulates symbiotic colonization traits of Vibrio fischeri and facilitates a beneficial association with an animal host. J. Bacteriol. 185, 7202–7212 (2003).
Whistler, C. A., Koropatnick, T. A., Pollack, A., McFall-Ngai, M. J. & Ruby, E. G. The GacA global regulator of Vibrio fischeri is required for normal host tissue responses that limit subsequent bacterial colonization. Cell. Microbiol. 9, 766–778 (2007).
Faraldo-Gomez, J. D. & Sansom, M. S. Acquisition of siderophores in Gram-negative bacteria. Nature Rev. Mol. Cell Biol. 4, 105–116 (2003).
Cowles, K. N., Cowles, C. E., Richards, G. R., Martens, E. C. & Goodrich-Blair, H. The global regulator Lrp contributes to mutualism, pathogenesis and phenotypic variation in the bacterium Xenorhabdus nematophila. Cell. Microbiol. 9, 1311–1323 (2007).
Graf, J. & Ruby, E. G. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc. Natl Acad. Sci. USA 95, 1818–1822 (1998).
Goetsch, M., Owen, H., Goldman, B. & Forst, S. Analysis of the PixA inclusion body protein of Xenorhabdus nematophila. J. Bacteriol. 188, 2706–2710 (2006).
Martens, E. C. et al. Xenorhabdus nematophila requires an intact iscRSUA–hscBA–fdx operon to colonize Steinernema carpocapsae nematodes. J. Bacteriol. 185, 3678–3682 (2003).
Dunn, A. K. & Stabb, E. V. The twin arginine translocation system contributes to symbiotic colonization of Euprymna scolopes by Vibrio fischeri. FEMS Microbiol. Lett. 279, 251–258 (2008).
Schaible, U. E. & Kaufmann, S. H. Iron and microbial infection. Nature Rev. Microbiol. 2, 946–953 (2004).
Watson, R. J., Joyce, S. A., Spencer, G. V. & Clarke, D. J. The exbD gene of Photorhabdus temperata is required for full virulence in insects and symbiosis with the nematode Heterorhabditis. Mol. Microbiol. 56, 763–773 (2005).
Crosa, J. H. & Walsh, C. T. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66, 223–249 (2002).
Davidson, S. K. & Stahl, D. A. Transmission of nephridial bacteria of the earthworm Eisenia fetida. Appl. Environ. Microbiol. 72, 769–775 (2006).
Davidson, S. K. & Stahl, D. A. Selective recruitment of bacteria during embryogenesis of an earthworm. ISME J. 2, 510–518 (2008).
Fraune, S. & Bosch, T. C. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl Acad. Sci. USA 104, 13146–13151 (2007).
Dale, C., Beeton, M., Harbison, C., Jones, T. & Pontes, M. Isolation, pure culture, and characterization of “Candidatus Arsenophonus arthropodicus,” an intracellular secondary endosymbiont from the hippoboscid louse fly Pseudolynchia canariensis. Appl. Environ. Microbiol. 72, 2997–3004 (2006).
Kikuchi, Y., Meng, X. Y. & Fukatsu, T. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae). Appl. Environ. Microbiol. 71, 4035–4043 (2005).
Kikuchi, Y., Hosokawa, T. & Fukatsu, T. Insect–microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 73, 4308–4316 (2007). Introduced an emerging experimental symbiosis system that had intriguing parallels with the squid– Vibrio association.
Ciche, T. A. & Sternberg, P. W. Postembryonic RNAi in Heterorhabditis bacteriophora: a nematode insect parasite and host for insect pathogenic symbionts. BMC Dev. Biol. 7, 101 (2007).
Acknowledgements
The author thanks M. McFall-Ngai for helpful discussions and ideas, and H. Goodrich-Blair, J. Graf and M. Mandel for reading parts of the manuscript. Support was provided by grants from the National Institutes of Health (grant number RR-12294) and the National Science Foundation (grant number IOB-0517007).
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Related links
Related links
DATABASES
Entrez Genome Project
FURTHER INFORMATION
Glossary
- Bioluminescence
-
The process by which some bacteria and other organisms produce light as the result of a chemical reaction. During symbiosis, this light can be used in behaviours such as counterillumination, in which the bioluminescence is used to eliminate the shadow of the host's silhouette.
- Gnotobiotic
-
An animal that is born under aseptic conditions and is exposed only to experimentally introduced microorganisms. Gnotobiotic animals are used to investigate the symbiotic relationship between an animal and one or more of the consortia of interacting microbial species that normally inhabit its body.
- Horizontal transfer
-
The process by which an animal or plant obtains its natural microbial constituents from the environment at each generation. By contrast, vertical transfer occurs when a young organism receives its microbiota from its parent, usually in or on the egg.
- Expressed sequence tag
-
(EST). One of a series of short nucleotide sequences which represent a pool of mRNAs that are expressed under a certain environmental or developmental condition. Libraries of ESTs can be used to identify gene transcripts in global expression studies.
- Two-component regulation system
-
A stimulus-response coupling mechanism that allows an organism to sense and respond to various changes in environmental conditions.
- Lipopolysaccharide
-
A major component of the outer membrane of Gram-negative bacteria. The immune systems of animals generally sense and react to the presence of lipopolysaccharide.
- Quorum sensing
-
A system by which bacteria respond to increased population density by coordinately controlling expression of a specific set of genes. By sensing the concentration of one of several continuously secreted signal molecules, including acyl homoserine lactones, peptides and autoinducer 2, the population can recognize when it reaches a 'quorum'.
- Auxotroph
-
An organism, or mutant derivative, which is unable to synthesize a particular compound (for example, an amino acid) that is required as a building block for its growth.
Rights and permissions
About this article
Cite this article
Ruby, E. Symbiotic conversations are revealed under genetic interrogation. Nat Rev Microbiol 6, 752–762 (2008). https://doi.org/10.1038/nrmicro1958
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1958
This article is cited by
-
The microbiome of the marine flatworm Macrostomum lignano provides fitness advantages and exhibits circadian rhythmicity
Communications Biology (2023)
-
RNA-Sequencing of Heterorhabditis nematodes to identify factors involved in symbiosis with Photorhabdus bacteria
BMC Genomics (2022)
-
Symbiont transmission in marine sponges: reproduction, development, and metamorphosis
BMC Biology (2022)
-
Impact of transit time on the reproductive capacity of Euprymna scolopes as a laboratory animal
Laboratory Animal Research (2022)
-
A lasting symbiosis: how the Hawaiian bobtail squid finds and keeps its bioluminescent bacterial partner
Nature Reviews Microbiology (2021)