Key Points
-
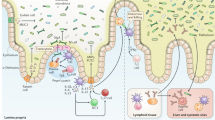
The human intestinal microbiota is dominated by anaerobic health-associated bacteria that are established and maintained in individuals through host-to-host transmission. The transmission process incorporates excretion from a host in faecal matter, survival and persistence in the external environment, and concludes with ingestion and subsequent colonization of a new host.
-
Studies of the routes of transmission of intestinal pathogens provide a useful framework to better understand intestinal commensal transmission. Both share some common transmission features, such as the use of the faecal–oral transmission route and similar survival mechanisms to persist in the external environment.
-
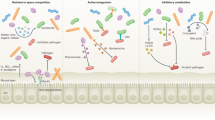
Environmental survival mechanisms that are used by the intestinal microbiota once expelled by a host include sporulation, aerotolerance and entering a viable but non-culturable (VBNC) dormancy state. For anaerobic bacteria, these mechanisms protect against harmful oxygen, and in the case of sporulation and VBNC states, can provide varying resistance against other environmental conditions, such as desiccation and a lack of nutrients.
-
Reservoirs are a source or a sink for bacteria during transmission. Other people in the community are the principal reservoirs of intestinal bacteria, but food, water, animals and the built environment may also facilitate transmission.
-
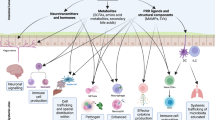
Transmission of commensal bacteria may be disrupted by human sanitation practices, through the use of antibiotics or a long-term change in diet, which can eliminate species within an individual thereby preventing their onward transmission. Direct interventions to restore a depleted microbiota, such as faecal microbiota transplantation (FMT), are effective in some cases.
-
A greater general awareness of the transmission of the commensal microbiota is facilitated by technological advances in different disciplines, including microbiology, bioinformatics and genomics. Fostering the transmission of commensal bacteria between people through the maintenance of a healthy lifestyle and the discerning use of antibiotics and sanitation processes may promote human health.
Abstract
Transmission of commensal intestinal bacteria between humans could promote health by establishing, maintaining and replenishing microbial diversity in the microbiota of an individual. Unlike pathogens, the routes of transmission for commensal bacteria remain unappreciated and poorly understood, despite the likely commonalities between both. Consequently, broad infection control measures that are designed to prevent pathogen transmission and infection, such as oversanitation and the overuse of antibiotics, may inadvertently affect human health by altering normal commensal transmission. In this Review, we discuss the mechanisms and factors that influence host-to-host transmission of the intestinal microbiota and examine how a better understanding of these processes will identify new approaches to nurture and restore transmission routes that are used by beneficial bacteria.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Whitman, W. B., Coleman, D. C. & Wiebe, W. J. Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. USA 95, 6578–6583 (1998).
Sekirov, I., Russell, S. L., Antunes, L. C. M. & Finlay, B. B. Gut microbiota in health and disease. Physiol. Rev. 90, 859–904 (2010).
Louis, P., Hold, G. L. & Flint, H. J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 12, 661–672 (2014).
Lawley, T. D. & Walker, A. W. Intestinal colonization resistance. Immunology 138, 1–11 (2013).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). This landmark study characterizes the microbiota across different human body sites.
Rajilic´-Stojanovic´, M. & de Vos, W. M. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 38, 996–1047 (2014). This study provides a detailed description and phylogeny of more than 1,000 cultured species from the intestinal microbiota.
Eckburg, P. B. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
Rolfe, R. D., Hentges, D. J., Campbell, B. J. & Barrett, J. T. Factors related to the oxygen tolerance of anaerobic bacteria. Appl. Environ. Microbiol. 36, 306–313 (1978).
Andriantsoanirina, V., Allano, S., Butel, M. J. & Aires, J. Tolerance of Bifidobacterium human isolates to bile, acid and oxygen. Anaerobe 21, 39–42 (2013).
Hug, L. A. et al. A new view of the tree of life. Nat. Microbiol. 1, 16048 (2016).
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R. & Gordon, J. I. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788 (2008).
Meehan, C. J. & Beiko, R. G. A. Phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol. Evol. 6, 703–713 (2014).
Surana, N. K. & Kasper, D. L. The yin yang of bacterial polysaccharides: lessons learned from B. fragilis PSA. Immunol. Rev. 245, 13–26 (2012).
Wexler, H. M. Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621 (2007).
Kamada, N., Chen, G. Y., Inohara, N. & Núñez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14, 685–690 (2013).
Chow, W.-H. et al. An inverse relation between cagA+ strains of Helicobacter pylori infection and risk of esophageal and gastric cardia adenocarcinoma. Cancer Res. 58, 588–590 (1998).
Petersen, C. & Round, J. L. Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 16, 1024–1033 (2014).
Koenig, J. E. et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl Acad. Sci. USA 108, 4578–4585 (2010).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Falkow, S. Who speaks for the microbes? Emerg. Infect. Dis. 4, 495–497 (1998).
Falkow, S. What is a pathogen? ASM News 63, 359–365 (1997).
Wexler, A. G. et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl Acad. Sci. USA 113, 3639–3644 (2016).
Duncan, S. H., Louis, P. & Flint, H. J. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 70, 5810–5817 (2004).
Rakoff-Nahoum, S., Foster, K. R. & Comstock, L. E. The evolution of cooperation within the gut microbiota. Nature 533, 255–259 (2016).
Turroni, F. et al. Expression of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in response to environmental gut conditions. FEMS Microbiol. Lett. 357, 23–33 (2014).
Neville, B. A. et al. Pro-inflammatory flagellin proteins of prevalent motile commensal bacteria are variably abundant in the intestinal microbiome of elderly humans. PLoS ONE 8, e68919 (2013).
He, M. et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat. Genet. 45, 109–113 (2013).
Harris, S. R. et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327, 469–474 (2010).
Chase-Topping, M., Gally, D., Low, C., Matthews, L. & Woolhouse, M. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nat. Rev. Microbiol. 6, 904–912 (2008).
Flo, T. H. et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432, 917–921 (2004).
Stecher, B. et al. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5, 2177–2189 (2007).
Rivera-Chávez, F. et al. Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe 19, 443–454 (2016).
Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2008).
Goodrich, J. K. et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014).
Turpin, W. et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat. Genet. 48, 1413–1417 (2016).
Pham, T. A. et al. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe 16, 504–516 (2014).
Bry, L., Falk, P. G., Midtvedt, T. & Gordon, J. I. A. Model of host–microbial interactions in an open mammalian ecosystem. Science 273, 1380–1383 (1996).
Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S. & Medzhitov, R. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004).
Brinkman, B. M. et al. Gut microbiota affects sensitivity to acute DSS-induced colitis independently of host genotype. Inflamm. Bowel Dis. 19, 2560–2567 (2013).
Cullen, T. W. et al. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 347, 170–175 (2015).
Sela, D. A. et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl Acad. Sci. USA 105, 18964–18969 (2008).
Charbonneau, M. R. et al. A microbial perspective of human developmental biology. Nature 535, 48–55 (2016).
Walker, A. W. et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 5, 220–230 (2010).
Flint, H. J., Scott, K. P., Louis, P. & Duncan, S. H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 9, 577–589 (2012).
Setlow, P. I will survive: DNA protection in bacterial spores. Trends Microbiol. 15, 172–180 (2007).
Errington, J. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1, 117–126 (2003).
Browne, H. P. et al. Culturing of 'unculturable' human microbiota reveals novel taxa and extensive sporulation. Nature 533, 543–546 (2016).
Atarashi, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013). This study shows that a selection of chloroform- resistant bacteria from human faeces can induce regulatory T cells in mice and have potential therapeutic properties through the alleviation of colitis in mice.
Janoir, C. et al. Adaptive strategies and pathogenesis of Clostridium difficile from in vivo transcriptomics. Infect. Immun. 81, 3757–3769 (2013).
Mora-Uribe, P. et al. Characterization of the adherence of Clostridium difficile spores: the integrity of the outermost layer affects adherence properties of spores of the epidemic strain R20291 to components of the intestinal mucosa. Front. Cell. Infect. Microbiol. 6, 99 (2016).
Duncan, S. H. et al. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int. J. Syst. Evol. Microbiol. 56, 2437–2441 (2006).
Flint, H. J., Duncan, S. H., Scott, K. P. & Louis, P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ. Microbiol. 9, 1101–1111 (2007).
Tally, F. P., Stewart, P. R., Sutter, V. L. & Rosenblatt, J. E. Oxygen tolerance of fresh clinical anaerobic bacteria. J. Clin. Microbiol. 1, 161–164 (1975).
Miller, R. A. & Britigan, B. E. Role of oxidants in microbial pathophysiology. Clin. Microbiol. Rev. 10, 1–18 (1997).
Khan, M. T. et al. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic–anoxic interphases. ISME J. 6, 1578–1585 (2012).
Donaldson, G. P., Lee, S. M. & Mazmanian, S. K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32 (2016).
Albenberg, L. et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 147, 1055–1063.e8 (2014).
Khan, M. T., van Dijl, J. M. & Harmsen, H. J. M. Antioxidants keep the potentially probiotic but highly oxygen-sensitive human gut bacterium Faecalibacterium prausnitzii alive at ambient air. PLoS ONE 9, e96097 (2014).
Li, L., Mendis, N., Trigui, H., Oliver, J. D. & Faucher, S. P. The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 5, 258 (2014).
Signoretto, C., Lleo, M. M., Tafi, M. C. & Canepari, P. Cell wall chemical composition of Enterococcus faecalis in the viable but nonculturable state. Appl. Environ. Microbiol. 66, 1953–1959 (2000).
Rittershaus, E. S., Baek, S.-H. & Sassetti, C. M. The normalcy of dormancy: common themes in microbial quiescence. Cell Host Microbe 13, 643–651 (2013).
Senoh, M. et al. Conversion of viable but nonculturable Vibrio cholerae to the culturable state by co-culture with eukaryotic cells. Microbiol. Immunol. 54, 502–507 (2010).
Amel, B. K., Amine, B. & Amina, B. Survival of Vibrio fluvialis in seawater under starvation conditions. Microbiol. Res. 163, 323–328 (2008).
Xu, H. S. et al. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb. Ecol. 8, 313–323 (1982).
Gupte, A. R., De Rezende, C. L. & Joseph, S. W. Induction and resuscitation of viable but nonculturable Salmonella enterica serovar Typhimurium DT104. Appl. Environ. Microbiol. 69, 6669–6675 (2003).
Dworkin, J. & Shah, I. M. Exit from dormancy in microbial organisms. Nat. Rev. Microbiol. 8, 890–896 (2010).
Backhed, F. et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703 (2015).
Dominguez-Bello, M. G. et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11971–11975 (2010).
Chu, D. M. et al. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 23, 314–326 (2017).
Blaser, M. J. & Falkow, S. What are the consequences of the disappearing human microbiota? Nat. Rev. Microbiol. 7, 887–894 (2009). This perspective proposes that changes in human ecology can compromise the composition of the human microbiota with associated negative effects on human health.
Arrieta, M. C., Stiemsma, L. T., Amenyogbe, N., Brown, E. M. & Finlay, B. The intestinal microbiome in early life: health and disease. Front. Immunol. 5, 427 (2014).
Schloss, P. D., Iverson, K. D., Petrosino, J. F. & Schloss, S. J. The dynamics of a family's gut microbiota reveal variations on a theme. Microbiome 2, 25 (2014).
Song, S. J. et al. Cohabiting family members share microbiota with one another and with their dogs. eLife 2, e00458 (2013).
Nayfach, S., Rodriguez-Mueller, B., Garud, N. & Pollard, K. S. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 26, 1612–1625 (2016). In this study, a SNP-based metagenomic pipeline is developed to demonstrate mother-to-infant vertical transmission, with early-colonizing bacteria being predominately non-spore forming and late-colonizing bacteria being predominately spore-forming, which indicates temporal environmental survival by spore-forming bacteria.
Faith, J. J., Colombel, J.-F. & Gordon, J. I. Identifying strains that contribute to complex diseases through the study of microbial inheritance. Proc. Natl Acad. Sci. USA 112, 633–640 (2015).
Tung, J. et al. Social networks predict gut microbiome composition in wild baboons. eLife 4, e05224 (2015).
Moeller, A. H. et al. Social behavior shapes the chimpanzee pan-microbiome. Sci. Adv. 2, e1500997 (2016).
De Filippo, C. et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl Acad. Sci. USA 107, 14691–14696 (2010).
Heikkila, M. P. & Saris, P. E. J. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J. Appl. Microbiol. 95, 471–478 (2003).
Fernandez, L. et al. The human milk microbiota: origin and potential roles in health and disease. Pharmacol. Res. 69, 1–10 (2013).
Derrien, M. & van Hylckama Vlieg, J. E. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 23, 354–366 (2015).
Jost, T., Lacroix, C., Braegger, C. P., Rochat, F. & Chassard, C. Vertical mother–neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 16, 2891–2904 (2014).
Vazquez-Torres, A. et al. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401, 804–808 (1999).
Martin, R. et al. The commensal microflora of human milk: new perspectives for food bacteriotherapy and probiotics. Trends Food Sci. Technol. 15, 121–127 (2004).
Lang, J. M., Eisen, J. A. & Zivkovic, A. M. The microbes we eat: abundance and taxonomy of microbes consumed in a day's worth of meals for three diet types. PeerJ 2, e659 (2014).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Leff, J. W. & Fierer, N. Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS ONE 8, e59310 (2013).
Brito, I. L. et al. Mobile genes in the human microbiome are structured from global to individual scales. Nature 535, 435–439 (2016).
Kalliomaki, M. et al. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet 357, 1076–1079 (2001).
Allen, S. J., Martinez, E. G., Gregorio, G. V. & Dans, L. F. Probiotics for treating acute infectious diarrhoea. Cochrane Database Syst. Rev. 11, CD003048 (2010).
Hill, C. et al. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514 (2014).
Malinen, E. et al. PCR-ELISAII: analysis of Bifidobacterium populations in human faecal samples from a consumption trial with Bifidobacterium lactis Bb-12 and a galacto-oligosaccharide preparation. Syst. Appl. Microbiol. 25, 249–258 (2002).
Charbonneau, D., Gibb, R. D. & Quigley, E. M. Fecal excretion of Bifidobacterium infantis 35624 and changes in fecal microbiota after eight weeks of oral supplementation with encapsulated probiotic. Gut Microbes 4, 201–211 (2013).
McNulty, N. P. et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci. Transl Med. 3, 106ra106 (2011).
Maldonado-Gómez, M. X. et al. Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe 20, 515–526 (2016).
Reid, G. et al. Responders and non-responders to probiotic interventions: how can we improve the odds? Gut Microbes 1, 200–204 (2010).
Sanders, M. E. et al. An update on the use and investigation of probiotics in health and disease. Gut 62, 787–796 (2013).
Natchu, U. C. & Bhatnagar, S. Diarrhoea in children: identifying the cause and burden. Lancet 382, 184–186 (2013).
Tallon, P., Magajna, B., Lofranco, C. & Leung, K. T. Microbial indicators of faecal contamination in water: a current perspective. Water Air Soil Poll. 166, 139–166 (2005).
Koskey, A. M. et al. Blautia and Prevotella sequences distinguish human and animal fecal pollution in Brazil surface waters. Environ. Microbiol. Rep. 6, 696–704 (2014).
Handl, S., Dowd, S. E., Garcia-Mazcorro, J. F., Steiner, J. M. & Suchodolski, J. S. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol. Ecol. 76, 301–310 (2011).
Hand, D., Wallis, C., Colyer, A. & Penn, C. W. Pyrosequencing the canine faecal microbiota: breadth and depth of biodiversity. PLoS ONE 8, e53115 (2013).
Lamendella, R., Domingo, J. W., Ghosh, S., Martinson, J. & Oerther, D. B. Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. 11, 103 (2011).
Ley, R. E. et al. Evolution of mammals and their gut microbes. Science 320, 1647–1651 (2008).
Moller-Stray, J. et al. Two outbreaks of diarrhoea in nurseries in Norway after farm visits, April to May 2009. Euro Surveill. 17, 20321 (2012).
Toro, M. et al. Whole-genome sequencing analysis of Salmonella enterica serovar Enteritidis isolates in Chile provides insights into possible transmission between gulls, poultry, and humans. Appl. Environ. Microbiol. 82, 6223–6232 (2016).
Knetsch, C. W. et al. Whole genome sequencing reveals potential spread of Clostridium difficile between humans and farm animals in the Netherlands, 2002 to 2011. Euro Surveill. 19, 20954 (2014).
Iovine, N. M. & Blaser, M. J. Antibiotics in animal feed and spread of resistant Campylobacter from poultry to humans. Emerg. Infect. Dis. 10, 1158–1189 (2004).
Gupta, A. et al. Antimicrobial resistance among Campylobacter strains, United States, 1997–2001. Emerg. Infect. Dis. 10, 1102–1109 (2004).
Dunn, R. R., Fierer, N., Henley, J. B., Leff, J. W. & Menninger, H. L. Home life: factors structuring the bacterial diversity found within and between homes. PLoS ONE 8, e64133 (2013).
Lax, S. et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048–1052 (2014).
Meadow, J. F. et al. Bacterial communities on classroom surfaces vary with human contact. Microbiome 2, 7 (2014).
Hsu, T. et al. Urban transit system microbial communities differ by surface type and interaction with humans and the environment. mSystems 1, e00018-16 (2016).
Kelley, S. T. & Gilbert, J. A. Studying the microbiology of the indoor environment. Genome Biol. 14, 202 (2013).
Kembel, S. W. et al. Architectural design influences the diversity and structure of the built environment microbiome. ISME J. 6, 1469–1479 (2012).
Barker, J. & Jones, M. V. The potential spread of infection caused by aerosol contamination of surfaces after flushing a domestic toilet. J. Appl. Microbiol. 99, 339–347 (2005).
Perkins, S. D., Mayfield, J., Fraser, V. & Angenent, L. T. Potentially pathogenic bacteria in shower water and air of a stem cell transplant unit. Appl. Environ. Microbiol. 75, 5363–5372 (2009).
Flores, G. E. et al. Microbial biogeography of public restroom surfaces. PLoS ONE 6, e28132 (2011).
Gibbons, S. M. et al. Ecological succession and viability of human-associated microbiota on restroom surfaces. Appl. Environ. Microbiol. 81, 765–773 (2015).
Snelling, A. M., Saville, T., Stevens, D. & Beggs, C. B. Comparative evaluation of the hygienic efficacy of an ultra-rapid hand dryer versus conventional warm air hand dryers. J. Appl. Microbiol. 110, 19–26 (2010).
Vaishampayan, P. et al. New perspectives on viable microbial communities in low-biomass cleanroom environments. ISME J. 7, 312–324 (2013).
Tsai, F. C. & Macher, J. M. Concentrations of airborne culturable bacteria in 100 large US office buildings from the BASE study. Indoor Air 15, 71–81 (2005).
David, L. A. et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 15, R89 (2014).
Korpela, K. et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 7, 10410 (2016).
Claesson, M. J. et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 488, 178–184 (2012).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Faith, J. J. et al. The long-term stability of the human gut microbiota. Science 341, 1237439 (2013).
Schloissnig, S. et al. Genomic variation landscape of the human gut microbiome. Nature 493, 45–50 (2012).
Costello, E. K., Stagaman, K., Dethlefsen, L., Bohannan, B. J. & Relman, D. A. The application of ecological theory toward an understanding of the human microbiome. Science 336, 1255–1262 (2012). This review uses ecological metacommunity theory to provide a framework to understand how the intestinal microbiota is assembled and to predict how it responds to disturbances.
Lee, S. M. et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501, 426–429 (2013).
Lawley, T. D. et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 8, e1002995 (2012).
Clemente, J. C. et al. The microbiome of uncontacted Amerindians. Sci. Adv. 1, e1500183 (2015).
Obregon-Tito, A. J. et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat. Commun. 6, 6505 (2015).
Sonnenburg, E. D. et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016).
Blaser, M. J. Antibiotic use and its consequences for the normal microbiome. Science 352, 544–545 (2016).
Martinez, I. et al. The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Rep. 11, 527–538 (2015).
Strachan, D. P. Hay fever, hygiene, and household size. BMJ 299, 1259–1260 (1989). Supporting the hygiene hypothesis, this study describes lower rates of hay fever and eczema in large families, which is associated with reduced hygiene among siblings in larger families.
Beasley, R. Worldwide variation in prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and atopic eczema: ISAAC. Lancet 351, 1225–1232 (1998).
Stiemsma, L., Reynolds, L., Turvey, S. & Finlay, B. The hygiene hypothesis: current perspectives and future therapies. Immunotargets Ther. 4, 143–157 (2015).
Gomez de Aguero, M. et al. The maternal microbiota drives early postnatal innate immune development. Science 351, 1296–1302 (2016).
Blaser, M. J. Who are we? Indigenous microbes and the ecology of human diseases. EMBO Rep. 7, 956–960 (2006).
Shaw, S. Y., Blanchard, J. F. & Bernstein, C. N. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am. J. Gastroenterol. 105, 2687–2692 (2010).
Dominguez-Bello, M. G. et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 22, 250–253 (2016).
van Nood, E. et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415 (2013). This study demonstrates the efficacy of faecal microbiota transplants to resolve C. difficile infection in a controlled clinical trial in humans.
Lagier, J.-C. et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 1, 16203 (2016).
Rettedal, E. A., Gumpert, H. & Sommer, M. O. A. Cultivation-based multiplex phenotyping of human gut microbiota allows targeted recovery of previously uncultured bacteria. Nat. Commun. 5, 4714 (2014).
Lau, J. T. et al. Capturing the diversity of the human gut microbiota through culture-enriched molecular profiling. Genome Med. 8, 72 (2016).
Buffie, C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2014). This study shows that a single bacterial species, Clostridium scindens , can promote resistance to infection with C. difficile in a mouse model through the synthesis of secondary bile acids that are inhibitory to this species, thus demonstrating the potential of defined bacteria-based therapies for the treatment of intestinal-associated disease.
Czaplewski, L. et al. Alternatives to antibiotics — a pipeline portfolio review. Lancet Infect. Dis. 16, 239–251 (2016).
Yee, A. L. & Gilbert, J. A. Microbiome. Is triclosan harming your microbiome? Science 353, 348–349 (2016).
Vandegrift, R. et al. Cleanliness in context: reconciling hygiene with a modern microbial perspective. Preprint at bioRxiv http://dx.doi.org/10.1101/095745 (2016).
Falkow, S. Molecular Koch's postulates applied to bacterial pathogenicity — a personal recollection 15 years later. Nat. Rev. Microbiol. 2, 67–72 (2004).
Parkhill, J. & Wren, B. W. Bacterial epidemiology and biology — lessons from genome sequencing. Genome Biol. 12, 230 (2011).
Brito, I. L. & Alm, E. J. Tracking strains in the microbiome: insights from metagenomics and models. Front. Microbiol. 7, 712 (2016).
Walker, A. W., Duncan, S. H., Louis, P. & Flint, H. J. Phylogeny, culturing, and metagenomics of the human gut microbiota. Trends Microbiol. 22, 267–274 (2014). This review highlights the importance of a multidisciplinary approach that combines metagenomic sequencing, bioinformatics and culturing to study the human intestinal microbiota.
Li, S. S. et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science 352, 586–589 (2016).
Subramanian, S. et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510, 417–421 (2014).
Lax, S., Nagler, C. R. & Gilbert, J. A. Our interface with the built environment: immunity and the indoor microbiota. Trends Immunol. 36, 121–123 (2015).
Forster, S. C. et al. HPMCD: the database of human microbial communities from metagenomic datasets and microbial reference genomes. Nucleic Acids Res. 44, D604–D609 (2015).
Rupnik, M., Wilcox, M. H. & Gerding, D. N. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7, 526–536 (2009).
Gough, E., Shaikh, H. & Manges, A. R. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin. Infect. Dis. 53, 994–1002 (2011).
Vyas, D., Aekka, A. & Vyas, A. Fecal transplant policy and legislation. World J. Gastroenterol. 21, 6–11 (2015).
Petrof, E. O. et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: 'RePOOPulating' the gut. Microbiome 1, 3 (2013).
Allegretti, J. R. & Hamilton, M. J. Restoring the gut microbiome for the treatment of inflammatory bowel diseases. World J. Gastroenterol. 20, 3468–3474 (2014).
Olle, B. Medicines from microbiota. Nat. Biotechnol. 31, 309–315 (2013).
Tvede, M. & Rask-Madsen, J. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet 1, 1156–1160 (1989).
Jalanka, J. et al. Long-term effects on luminal and mucosal microbiota and commonly acquired taxa in faecal microbiota transplantation for recurrent Clostridium difficile infection. BMC Med. 14, 155 (2016).
Acknowledgements
The authors thank A. Zhu for critical reading of the manuscript during preparation. H.P.B. is funded through a Medical Research Council (MRC) grant (PF451), B.A.N. is funded through a Wellcome Trust grant (098051), S.C.F. is funded through an Australian National Health and Medical Research Council (NHMRC) grant (1091097). Work in the Lawley laboratory is supported through a MRC grant (PF451) and a Wellcome Trust grant (098051).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information Table 1
Aerotolerance of non-spore forming intestinal bacteria. (PDF 195 kb)
Glossary
- Peritoneum
-
A membrane that lines the abdominal cavity and provides a supportive role to internal body organs, including those of the gastrointestinal tract.
- Pathobionts
-
Members of the commensal microbiota that may become pathogenic under certain circumstances.
- Facultative anaerobic bacteria
-
Species that can grow and survive in aerobic and anaerobic conditions.
- Infectious dose
-
The minimum number of bacteria required to cause an infection in a host.
- Colonization resistance
-
The capacity of the resident microbiota to prevent the establishment of new species within the community, particularly the establishment of pathogens. Colonization resistance is a feature of a stable health-associated microbiota.
- Homeostasis
-
A stable state. The overall maintenance of precise conditions in the microbiota that promote colonization resistance, even when subjected to external perturbations or stresses.
- Colonizing dose
-
The minimum number of bacteria required to stably colonize a new host.
- Super-shedding state
-
A host state, typically associated with infection by pathogens, that results in the release of numerous bacteria or spores into the external environment.
- Antimicrobial peptides
-
A diverse range of proteins that are secreted by the host as a defence mechanism against pathogens or by microorganisms to target other microorganisms in close proximity.
- Siderophores
-
Low-molecular-weight, iron-chelating agents that are secreted by bacteria and fungi to acquire iron from the surrounding environment.
- Dysbiosis
-
A low-diversity microbiota with reduced colonization resistance that is typically associated with inflammation and outgrowth of facultative anaerobic Proteobacteria and pathogens.
- Fucosylated
-
The attachment of a fucose molecule to a protein. Fucosylation of epithelial cells by the host can provide a protective role through the subsequent recruitment of commensal bacteria.
- Toll-like receptors
-
A family of transmembrane protein receptors, characterized by the presence of a Toll and interleukin-1 receptor (TIR) domain, that recognize specific microorganism-associated molecular patterns and initiate an immune response.
- Nucleotide-binding oligomerization domain-like receptors
-
(NOD-like receptors). A family of intracellular protein receptors that recognize microorganism-associated molecular patterns and initiate an immune response.
- Desiccation
-
The process of liquid removal or drying out, which is usually deleterious to a bacterial cell.
- Vegetative cell
-
The form of a bacterial cell that reproduces through binary fission.
- Aerobic bacteria
-
Species that can only grow and survive in the presence of oxygen.
- Obligate anaerobic bacteria
-
Species that can only grow and survive in the absence of oxygen.
- Flavin–thiol electron shuttle
-
A process that involves the transfer of electrons to oxygen through riboflavin and thiol, which enables survival and growth in the presence of oxygen.
- Social grooming
-
Cleaning and grooming carried out by animals, particularly primates, on other individuals in their community, which has hygienic and social roles.
- Horizontal gene transfer
-
The transfer of genetic material between different strains or species that occurs independently of vertical transmission during replication.
- Probiotics
-
Live microorganisms that when administered in adequate amounts confer a health benefit to the host.
- Indicator microorganisms
-
Microorganisms that are used to assay hygiene levels of foods or water, in which the quantity of the microorganisms present is inversely related to the quality or hygiene level of the product being tested.
- Undernutrition
-
A situation whereby an individual does not consume enough nutrients, which can have adverse effects on health.
- α-diversity
-
The ecological diversity at a single site, as measured by the number of different species and their abundance.
- β-diversity
-
A measure of the difference in ecological diversity between different sites.
- Functional foods
-
Foods that contain additional elements to promote health, such as probiotics, prebiotics, vitamins or minerals.
Rights and permissions
About this article
Cite this article
Browne, H., Neville, B., Forster, S. et al. Transmission of the gut microbiota: spreading of health. Nat Rev Microbiol 15, 531–543 (2017). https://doi.org/10.1038/nrmicro.2017.50
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2017.50
This article is cited by
-
Uncovering the microbiome landscape in sashimi delicacies
Scientific Reports (2024)
-
Core gut microbes Cloacibacterium and Aeromonas associated with different gastropod species could be persistently transmitted across multiple generations
Microbiome (2023)
-
Transmission of Alzheimer’s disease-associated microbiota dysbiosis and its impact on cognitive function: evidence from mice and patients
Molecular Psychiatry (2023)
-
Elucidating the transmission landscape of the human microbiome
Nature Reviews Gastroenterology & Hepatology (2023)
-
The person-to-person transmission landscape of the gut and oral microbiomes
Nature (2023)