Key Points
-
At the start of each ubiquitin-like protein (UBL) cascade is an E1 enzyme, which activates the UBL and then directs the UBL to downstream pathways. UBL proteins and E1 (or activating) enzymes have their origins in prokaryotic biosynthetic pathways.
-
In humans, eight E1 enzymes are known to initiate UBL conjugation. We refer to E1s for ubiquitin (UBA1 and UBA6 (also known as UBE1L2)), SUMO (SAE1–UBA2), NEDD8 (NAE1–UBA3) and ISG15 (UBA7) as canonical, owing to their related domain structures and enzymatic mechanisms, and to the more divergent E1s for URM1 (UBA4), UFM1 (UBA5) and ATG12 and ATG8 isoforms (ATG7) as non-canonical. (ATG is autophagy-related protein; ISG15 is interferon-stimulated gene 15; NAE1 is NEDD8-activating enzyme 1; SAE1 is SUMO-activating enzyme 1; UBA is ubiquitin-activating enzyme; UFM1 is ubiquitin-fold modifier 1; URM1 is ubiquitin-related modifier 1).
-
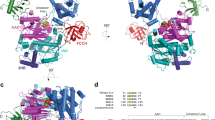
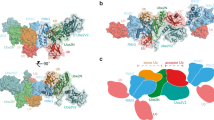
Canonical E1 structures all have an adenylation domain that resembles prokaryotic ancestors, and two domains that are specific to canonical E1s: a catalytic Cys domain that contains the Cys that is involved in the E1∼UBL thioester linkage and a carboxyl-terminal ubiquitin-fold domain that resembles ubiquitin and that binds E2.
-
In addition to their chemical roles in initiating UBL conjugation cascades, E1s also establish specificity, by matching a particular UBL with only cognate E2s. Rules for how this specificity is achieved are only beginning to emerge, but it is clear that specificity is achieved at multiple levels.
-
A particularly intriguing non-canonical E1 is UBA4 (MOCS3 in humans). Recent data indicate that UBA4 initiates a sulphur transfer pathway, in a manner related to bacterial E1-like enzymes.
-
E1s are unique in the UBL conjugating pathway in that they are the only components that use ATP, and this property is being exploited to develop selective inhibitors. The potential for the use of such inhibitors in therapy is high, given the links seen between components of UBL cascades and human disease.
Abstract
Attachment of ubiquitin or ubiquitin-like proteins (known as UBLs) to their targets through multienzyme cascades is a central mechanism to modulate protein functions. This process is initiated by a family of mechanistically and structurally related E1 (or activating) enzymes. These activate UBLs through carboxy-terminal adenylation and thiol transfer, and coordinate the use of UBLs in specific downstream pathways by charging cognate E2 (or conjugating) enzymes, which then interact with the downstream ubiquitylation machinery to coordinate the modification of the target. A broad understanding of how E1 enzymes activate UBLs and how they selectively coordinate UBLs with downstream function has come from enzymatic, structural and genetic studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
09 April 2009
In the main text, 'inorganic phosphate' in the following two sentences should read 'inorganic pyrophosphate': "The basic side chains of MoaD or ThiS would stabilize the developing negative charge from the ensuing pentacovalent phosphate intermediate, allowing the generation of MoaD∼adenylate or ThiS∼adenylate and inorganic phosphate." And: "Progression of the cascade is driven by the release of the small molecule products inorganic phosphate and AMP in the first and second steps, respectively56,57,58,62,63."
References
Ciechanover, A. Intracellular protein degradation: from a vague idea, through the lysosome and the ubiquitin–proteasome system, and onto human diseases and drug targeting (Nobel lecture). Angew. Chem. Int. Edn Engl. 44, 5944–5967 (2005).
Goldberg, A. L. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem. Soc. Trans. 35, 12–17 (2007).
Hershko, A., Heller, H., Elias, S. & Ciechanover, A. Components of ubiquitin–protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 258, 8206–8214 (1983). Describes the chromatographic separation of E1, E2 and E3 activities and demonstrates reconstitution of the first ubiquitin ligase reaction.
Hochstrasser, M. Lingering mysteries of ubiquitin-chain assembly. Cell 124, 27–34 (2006).
Pickart, C. M. & Fushman, D. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610–616 (2004).
Hicke, L., Schubert, H. L. & Hill, C. P. Ubiquitin-binding domains. Nature Rev. Mol. Cell Biol. 6, 610–621 (2005).
Hochstrasser, M. Ubiquitin signalling: what's in a chain? Nature Cell Biol. 6, 571–572 (2004).
Hurley, J. H., Lee, S. & Prag, G. Ubiquitin-binding domains. Biochem. J. 399, 361–372 (2006).
Harper, J. W. & Schulman, B. A. Structural complexity in ubiquitin recognition. Cell 124, 1133–1136 (2006).
Ciechanover, A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nature Rev. Mol. Cell Biol. 6, 79–87 (2005).
Hershko, A. The ubiquitin system for protein degradation and some of its roles in the control of the cell-division cycle (Nobel lecture). Angew. Chem. Int. Edn Engl. 44, 5932–5943 (2005).
Hochstrasser, M. Biochemistry. All in the ubiquitin family. Science 289, 563–564 (2000).
Hochstrasser, M. Evolution and function of ubiquitin-like protein-conjugation systems. Nature Cell Biol. 2, E153–E157 (2000).
Kerscher, O., Felberbaum, R. & Hochstrasser, M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 (2006).
Pickart, C. M. & Eddins, M. J. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 (2004).
Varshavsky, A. Regulated protein degradation. Trends Biochem. Sci. 30, 283–286 (2005).
Hershko, A., Ciechanover, A. & Varshavsky, A. The ubiquitin system. Nature Med. 6, 1073–1081 (2000).
Yeh, E. T., Gong, L. & Kamitani, T. Ubiquitin-like proteins: new wines in new bottles. Gene 248, 1–14 (2000).
Pan, Z. Q., Kentsis, A., Dias, D. C., Yamoah, K. & Wu, K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23, 1985–1997 (2004).
Yamoah, K. et al. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1's C-terminal tail. Proc. Natl Acad. Sci. USA 105, 12230–12235 (2008).
Duda, D. M. et al. Structural insights into NEDD8 activation of cullin–RING ligases: conformational control of conjugation. Cell 134, 995–1006 (2008).
Saha, A. & Deshaies, R. J. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32, 21–31 (2008).
Geiss-Friedlander, R. & Melchior, F. Concepts in sumoylation: a decade on. Nature Rev. Mol. Cell Biol. 8, 947–956 (2007).
Wang, C., Xi, J., Begley, T. P. & Nicholson, L. K. Solution structure of ThiS and implications for the evolutionary roots of ubiquitin. Nature Struct. Biol. 8, 47–51 (2001).
Lake, M. W., Wuebbens, M. M., Rajagopalan, K. V. & Schindelin, H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB–MoaD complex. Nature 414, 325–329 (2001). Describes the structure of the MoeB–MoaD complex and reveals the mechanism of MoaD adenylation. This is the only structure to date of an E1-like enzyme with an adenylate.
Taylor, S. V. et al. Thiamin biosynthesis in Escherichia coli. Identification of this thiocarboxylate as the immediate sulfur donor in the thiazole formation. J. Biol. Chem. 273, 16555–16560 (1998).
Leimkuhler, S., Wuebbens, M. M. & Rajagopalan, K. V. Characterization of Escherichia coli MoeB and its involvement in the activation of molybdopterin synthase for the biosynthesis of the molybdenum cofactor. J. Biol. Chem. 276, 34695–34701 (2001).
Duda, D. M., Walden, H., Sfondouris, J. & Schulman, B. A. Structural analysis of Escherichia coli ThiF. J. Mol. Biol. 349, 774–786 (2005).
Lehmann, C., Begley, T. P. & Ealick, S. E. Structure of the Escherichia coli ThiS–ThiF complex, a key component of the sulfur transfer system in thiamin biosynthesis. Biochemistry 45, 11–19 (2006).
Sloper-Mould, K. E., Jemc, J. C., Pickart, C. M. & Hicke, L. Distinct functional surface regions on ubiquitin. J. Biol. Chem. 276, 30483–30489 (2001).
Roush, R. F., Nolan, E. M., Lohr, F. & Walsh, C. T. Maturation of an Escherichia coli ribosomal peptide antibiotic by ATP-consuming N–P bond formation in microcin C7. J. Am. Chem. Soc. 130, 3603–3609 (2008).
Pearce, M. J., Mintseris, J., Ferreyra, J., Gygi, S. P. & Darwin, K. H. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 322, 1104–1107 (2008).
McGrath, J. P., Jentsch, S. & Varshavsky, A. UBA 1: an essential yeast gene encoding ubiquitin-activating enzyme. EMBO J. 10, 227–236 (1991).
Handley, P. M., Mueckler, M., Siegel, N. R., Ciechanover, A. & Schwartz, A. L. Molecular cloning, sequence, and tissue distribution of the human ubiquitin-activating enzyme E1. Proc. Natl Acad. Sci. USA 88, 258–262 (1991).
Hatfield, P. M., Callis, J. & Vierstra, R. D. Cloning of ubiquitin activating enzyme from wheat and expression of a functional protein in Escherichia coli. J. Biol. Chem. 265, 15813–15817 (1990).
Liakopoulos, D., Doenges, G., Matuschewski, K. & Jentsch, S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 17, 2208–2214 (1998).
Gong, L. & Yeh, E. T. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J. Biol. Chem. 274, 12036–12042 (1999).
Desterro, J. M., Rodriguez, M. S., Kemp, G. D. & Hay, R. T. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 274, 10618–10624 (1999).
Gong, L., Li, B., Millas, S. & Yeh, E. T. Molecular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex. FEBS Lett. 448, 185–189 (1999).
Johnson, E. S., Schwienhorst, I., Dohmen, R. J. & Blobel, G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16, 5509–5519 (1997).
Yuan, W. & Krug, R. M. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20, 362–371 (2001).
Furukawa, K., Mizushima, N., Noda, T. & Ohsumi, Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J. Biol. Chem. 275, 7462–7465 (2000).
Komatsu, M. et al. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 23, 1977–1986 (2004).
Jin, J., Li, X., Gygi, S. P. & Harper, J. W. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 447, 1135–1138 (2007). Together with references 45 and 46, this paper identifies UBA6 as a second activating enzyme for ubiquitin, and identifies USE1 as a specific E2 conjugating enzyme for UBA6.
Pelzer, C. et al. UBE1L2, a novel E1 enzyme specific for ubiquitin. J. Biol. Chem. 282, 23010–23014 (2007).
Chiu, Y. H., Sun, Q. & Chen, Z. J. E1-L2 activates both ubiquitin and FAT10. Mol. Cell 27, 1014–1023 (2007). Identifies UBA6 as a candidate E1 for FAT10.
Mizushima, N. et al. A protein conjugation system essential for autophagy. Nature 395, 395–398 (1998).
Huang, D. T., Walden, H., Duda, D. & Schulman, B. A. Ubiquitin-like protein activation. Oncogene 23, 1958–1971 (2004).
Walden, H., Podgorski, M. S. & Schulman, B. A. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature 422, 330–334 (2003). Describes the first crystal structure of the canonical E1 protein complex responsible for activating NEDD8, and defines the domain structure of canonical E1s.
Lois, L. M. & Lima, C. D. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 24, 439–451 (2005). Describes the first structure of the heterodimeric SUMO activating enzyme bound to SUMO.
Lee, I. & Schindelin, H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell 134, 268–278 (2008). Describes the first structure of an activating enzyme for ubiquitin in complex with ubiquitin.
Komatsu, M. et al. The C-terminal region of an Apg7p/Cvt2p is required for homodimerization and is essential for its E1 activity and E1–E2 complex formation. J. Biol. Chem. 276, 9846–9854 (2001).
Tanida, I., Tanida-Miyake, E., Ueno, T. & Kominami, E. The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J. Biol. Chem. 276, 1701–1706 (2001).
Schmitz, J. et al. The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry 47, 6479–6489 (2008).
Ichimura, Y. et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 (2000).
Haas, A. L. & Rose, I. A. The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J. Biol. Chem. 257, 10329–10337 (1982).
Haas, A. L., Warms, J. V., Hershko, A. & Rose, I. A. Ubiquitin-activating enzyme. Mechanism and role in protein–ubiquitin conjugation. J. Biol. Chem. 257, 2543–2548 (1982). Demonstrates that UBA1 becomes doubly loaded with ubiquitin, with one molecule in a thioester with the catalytic Cys of UBA1 and the second ubiquitin as an adenylate.
Haas, A. L., Warms, J. V. & Rose, I. A. Ubiquitin adenylate: structure and role in ubiquitin activation. Biochemistry 22, 4388–4394 (1983).
Ciechanover, A., Heller, H., Katz-Etzion, R. & Hershko, A. Activation of the heat-stable polypeptide of the ATP-dependent proteolytic system. Proc. Natl Acad. Sci. USA 78, 761–765 (1981). Suggests a two step mechanism for ubiquitin activation, the first involving adenylation and the second involving thioester formation.
Ciechanover, A., Elias, S., Heller, H. & Hershko, A. “Covalent affinity” purification of ubiquitin-activating enzyme. J. Biol. Chem. 257, 2537–2542 (1982). Describes the purification of E1 ubiquitin activating enzyme, UBA1, which used the thioester as a tool for recovery of active enzyme. The ability to purify active UBA1 revolutionized the field.
Pickart, C. M. & Rose, I. A. Functional heterogeneity of ubiquitin carrier proteins. J. Biol. Chem. 260, 1573–1581 (1985). Demonstrated the existence of a family of E2 conjugating enzymes that are charged by UBA1, foreshadowing the discovery that distinct E2s are involved in distinct biological pathways.
Haas, A. L. & Bright, P. M. The resolution and characterization of putative ubiquitin carrier protein isozymes from rabbit reticulocytes. J. Biol. Chem. 263, 13258–13267 (1988).
Haas, A. L., Bright, P. M. & Jackson, V. E. Functional diversity among putative E2 isozymes in the mechanism of ubiquitin–histone ligation. J. Biol. Chem. 263, 13268–13275 (1988).
Pickart, C. M., Kasperek, E. M., Beal, R. & Kim, A. Substrate properties of site-specific mutant ubiquitin protein (G76A) reveal unexpected mechanistic features of ubiquitin-activating enzyme (E1). J. Biol. Chem. 269, 7115–7123 (1994).
Tokgoz, Z., Bohnsack, R. N. & Haas, A. L. Pleiotropic effects of ATP..Mg2+ binding in the catalytic cycle of ubiquitin-activating enzyme. J. Biol. Chem. 281, 14729–14737 (2006).
Bohnsack, R. N. & Haas, A. L. Conservation in the mechanism of Nedd8 activation by the human AppBp1–Uba3 heterodimer. J. Biol. Chem. 278, 26823–26830 (2003).
Walden, H. et al. The structure of the APPBP1–UBA3–NEDD8–ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol. Cell 12, 1427–1437 (2003).
Huang, D. T. et al. Basis for a ubiquitin-like protein thioester switch toggling E1–E2 affinity. Nature 445, 394–398 (2007). Reports the first structure of a doubly loaded canonical E1 enzyme, and defines a thioester switch responsible for promoting transfer of the ubiquitin like protein NEDD8 to its E2, UBC12.
Zhao, C. et al. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-α/β-induced ubiquitin-like protein. Proc. Natl Acad. Sci. USA 101, 7578–7582 (2004).
Huang, D. T. et al. A unique E1–E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nature Struct. Mol. Biol. 11, 927–935 (2004).
Huang, D. T. et al. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1. Mol. Cell 17, 341–350 (2005).
Souphron, J. et al. Structural dissection of a gating mechanism preventing misactivation of ubiquitin by NEDD8's E1. Biochemistry 47, 8961–8969 (2008).
Wilkinson, K. D. et al. A specific inhibitor of the ubiquitin activating enzyme: synthesis and characterization of adenosyl-phospho-ubiquitinol, a nonhydrolyzable ubiquitin adenylate analogue. Biochemistry 29, 7373–7380 (1990).
Madden, M. M. et al. Substrate properties of ubiquitin carboxyl-terminally derived peptide probes for protein ubiquitination. Biochemistry 47, 3636–3644 (2008).
Szczepanowski, R. H., Filipek, R. & Bochtler, M. Crystal structure of a fragment of mouse ubiquitin-activating enzyme. J. Biol. Chem. 280, 22006–22011 (2005).
Sullivan, M. L. & Vierstra, R. D. Cloning of a 16-kDa ubiquitin carrier protein from wheat and Arabidopsis thaliana. Identification of functional domains by in vitro mutagenesis. J. Biol. Chem. 266, 23878–23885 (1991).
Bencsath, K. P., Podgorski, M. S., Pagala, V. R., Slaughter, C. A. & Schulman, B. A. Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J. Biol. Chem. 277, 47938–47945 (2002).
Durfee, L. A., Kelley, M. L. & Huibregtse, J. M. The basis for selective E1–E2 interactions in the ISG15 conjugation system. J. Biol. Chem. 283, 23895–23902 (2008).
Lee, T. V. et al. The E1 ubiquitin-activating enzyme Uba1 in Drosophila controls apoptosis autonomously and tissue growth non-autonomously. Development 135, 43–52 (2008).
Eletr, Z. M., Huang, D. T., Duda, D. M., Schulman, B. A. & Kuhlman, B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nature Struct. Mol. Biol. 12, 933–934 (2005).
Reverter, D. & Lima, C. D. Insights into E3 ligase activity revealed by a SUMO–RanGAP1–Ubc9–Nup358 complex. Nature 435, 687–692 (2005).
Rabut, G. & Peter, M. Function and regulation of protein neddylation. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep. 9, 969–976 (2008).
Tatham, M. H. et al. Role of an N-terminal site of Ubc9 in SUMO-1, -2, and -3 binding and conjugation. Biochemistry 42, 9959–9969 (2003).
Whitby, F. G., Xia, G., Pickart, C. M. & Hill, C. P. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J. Biol. Chem. 273, 34983–34991 (1998).
Wang, J. et al. The intrinsic affinity between E2 and the Cys domain of E1 in ubiquitin-like modifications. Mol. Cell 27, 228–237 (2007).
Haas, A. L. Structural insights into early events in the conjugation of ubiquitin and ubiquitin-like proteins. Mol. Cell 27, 174–175 (2007).
Huang, D. T., Zhuang, M., Ayrault, O. & Schulman, B. A. Identification of conjugation specificity determinants unmasks vestigial preference for ubiquitin within the NEDD8 E2. Nature Struct. Mol. Biol. 15, 280–287 (2008).
Finley, D., Ciechanover, A. & Varshavsky, A. Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell 37, 43–55 (1984). Identifies UBA1 as the protein mutated in the ts85 cell line and demonstrates that UBA1 is responsible for the vast majority of ubiquitin conjugation in mammalian cells.
Ciechanover, A., Finley, D. & Varshavsky, A. Mammalian cell cycle mutant defective in intracellular protein degradation and ubiquitin–protein conjugation. Prog. Clin. Biol. Res. 180, 17–31 (1985).
Schlabach, M. R. et al. Cancer proliferation gene discovery through functional genomics. Science 319, 620–624 (2008).
Strous, G. J., van Kerkhof, P., Govers, R., Ciechanover, A. & Schwartz, A. L. The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. EMBO J. 15, 3806–3812 (1996).
Rocca, A., Lamaze, C., Subtil, A. & Dautry-Varsat, A. Involvement of the ubiquitin/proteasome system in sorting of the interleukin 2 receptor β chain to late endocytic compartments. Mol. Biol. Cell 12, 1293–1301 (2001).
Swanson, R. & Hochstrasser, M. A viable ubiquitin-activating enzyme mutant for evaluating ubiquitin system function in Saccharomyces cerevisiae. FEBS Lett. 477, 193–198 (2000).
Ghaboosi, N. & Deshaies, R. J. A conditional yeast E1 mutant blocks the ubiquitin–proteasome pathway and reveals a role for ubiquitin conjugates in targeting Rad23 to the proteasome. Mol. Biol. Cell 18, 1953–1963 (2007).
Kulkarni, M. & Smith, H. E. E1 ubiquitin-activating enzyme UBA-1 plays multiple roles throughout C. elegans development. PLoS Genet. 4, e1000131 (2008).
Pfleger, C. M., Harvey, K. F., Yan, H. & Hariharan, I. K. Mutation of the gene encoding the ubiquitin activating enzyme Uba1 causes tissue overgrowth in Drosophila. Fly (Austin) 1, 95–105 (2007).
Nollen, E. A. et al. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc. Natl Acad. Sci. USA 101, 6403–6408 (2004).
Ramser, J. et al. Rare missense and synonymous variants in UBE1 are associated with X-linked infantile spinal muscular atrophy. Am. J. Hum. Genet. 82, 188–193 (2008).
Stephen, A. G., Trausch-Azar, J. S., Ciechanover, A. & Schwartz, A. L. The ubiquitin-activating enzyme E1 is phosphorylated and localized to the nucleus in a cell cycle-dependent manner. J. Biol. Chem. 271, 15608–15614 (1996).
Nouspikel, T. & Hanawalt, P. C. Impaired nucleotide excision repair upon macrophage differentiation is corrected by E1 ubiquitin-activating enzyme. Proc. Natl Acad. Sci. USA 103, 16188–16193 (2006).
Grenfell, S. J., Trausch-Azar, J. S., Handley-Gearhart, P. M., Ciechanover, A. & Schwartz, A. L. Nuclear localization of the ubiquitin-activating enzyme, E1, is cell-cycle-dependent. Biochem. J. 300, 701–708 (1994).
Dohmen, R. J. et al. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J. Biol. Chem. 270, 18099–18109 (1995).
Bossis, G. & Melchior, F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol. Cell 21, 349–357 (2006).
Boggio, R., Colombo, R., Hay, R. T., Draetta, G. F. & Chiocca, S. A mechanism for inhibiting the SUMO pathway. Mol. Cell 16, 549–561 (2004).
Boggio, R., Passafaro, A. & Chiocca, S. Targeting SUMO E1 to ubiquitin ligases: a viral strategy to counteract sumoylation. J. Biol. Chem. 282, 15376–15382 (2007).
Handeli, S. & Weintraub, H. The ts41 mutation in Chinese hamster cells leads to successive S phases in the absence of intervening G2, M, and G1. Cell 71, 599–611 (1992).
Chen, Y., McPhie, D. L., Hirschberg, J. & Neve, R. L. The amyloid precursor protein-binding protein APP-BP1 drives the cell cycle through the S–M checkpoint and causes apoptosis in neurons. J. Biol. Chem. 275, 8929–8935 (2000).
Kumar, A. et al. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. EMBO J. 26, 4457–4466 (2007).
Goehring, A. S., Rivers, D. M. & Sprague, G. F. Jr. Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryot. Cell 2, 930–936 (2003).
Schlieker, C., Van der Veen, A. G., Damon, J. R., Spooner, E. & Ploegh, H. L. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc. Natl Acad. Sci. USA 105, 18255–18260 (2008).
Leidel, S. et al. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 458, 228–232 (2009).
Nakai, Y., Nakai, M. & Hayashi, H. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J. Biol. Chem. 283, 27469–27476 (2008).
Huang, B., Lu, J. & Bystrom, A. S. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA 14, 2183–2194 (2008).
Tanida, I. et al. Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell 10, 1367–1379 (1999).
Yamada, Y. et al. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J. Biol. Chem. 282, 8036–8043 (2007).
Mizushima, T. et al. Crystal structure of Ufc1, the Ufm1-conjugating enzyme. Biochem. Biophys. Res. Commun. 362, 1079–1084 (2007).
Sekizawa, R. et al. Panepophenanthrin, from a mushroom strain, a novel inhibitor of the ubiquitin-activating enzyme. J. Nat. Prod. 65, 1491–1493 (2002).
Yang, Y. et al. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 67, 9472–9481 (2007).
Tong, H., Hateboer, G., Perrakis, A., Bernards, R. & Sixma, T. K. Crystal structure of murine/human Ubc9 provides insight into the variability of the ubiquitin-conjugating system. J. Biol. Chem. 272, 21381–21387 (1997).
Worthylake, D. K., Prakash, S., Prakash, L. & Hill, C. P. Crystal structure of the Saccharomyces cerevisiae ubiquitin-conjugating enzyme Rad6 at 2.6 Å resolution. J. Biol. Chem. 273, 6271–6276 (1998).
Soucy, T. A. et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature (in the press).
Huang, D.T. et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell. 33, 483–495 (2009).
Acknowledgements
We thank the National Institutes of Health (J.W.H. and B.A.S.) and Millennium Pharmaceuticals, Inc. (J.W.H.) for funding our work on E1 enzymes and C. Regni for assistance with figure 2. B.A.S. is an Investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
Brenda A. Schulman is a consultant for Millennium Pharmaceuticals.
J. Wade Harper is also a consultant for and receives grant support from Millennium Pharmaceuticals.
Related links
Related links
DATABASES
Protein Data Bank
FURTHER INFORMATION
Glossary
- Adenylation
-
The synthesis of a phosphodiester bond between a hydroxyl group and the phosphate group of AMP. In the case of ubiquitin-like protein (UBL) conjugation cascades, the hydroxyl is from the carboxyl terminus of the UBL.
- Thioester bond
-
Covalent linkage of a sulphur with an acyl group. In the case of ubiquitin-like protein (UBL) cascades, the Cys sulphur of an enzyme is linked to the terminal carbon of a UBL.
- 26S proteasome
-
A large multisubunit protease complex that selectively degrades polyubiquitylated proteins. It contains a 20S particle that carries the catalytic activity and two regulatory 19S particles.
- UBL fold
-
A structural motif, also known as βGRASP fold, that has a domain with a structure of two β-sheets, followed by an α-helix and two additional β-sheets.
- Rhodanese homology domain
-
A domain that shares sequence and structural homology to rhodanese, a mitochondrial enzyme that catalyses Cys-mediated sulphur transfer reactions.
- PP loop ATPase domain
-
A structural motif found in a family of conserved ATP-binding proteins that serve as tRNA-modifying enzymes and activate the tRNA through adenylation.
- Elongator complex
-
An enzyme complex that replaces the hydrogen atom on position 5 of uridine at nucleotide 34 of tRNAs by a methoxycarbonylmethyl group.
- Thiouridinylation
-
An enzymatic process in which the 2' oxygen of uridine in the anticodon of a tRNA is replaced by sulphur.
Rights and permissions
About this article
Cite this article
Schulman, B., Wade Harper, J. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol 10, 319–331 (2009). https://doi.org/10.1038/nrm2673
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm2673
This article is cited by
-
Deciphering the role of neddylation in tumor microenvironment modulation: common outcome of multiple signaling pathways
Biomarker Research (2024)
-
Lighting up kinase contacts in situ
Nature Chemical Biology (2024)
-
The Vitis yeshanensis U-box E3 ubiquitin ligase VyPUB21 enhances resistance to powdery mildew by targeting degradation of NIM1-interacting (NIMIN) protein
Plant Cell Reports (2024)
-
Ubiquitination Process Mediates Prostate Cancer Development and Metastasis through Multiple Mechanisms
Cell Biochemistry and Biophysics (2024)
-
Neutron-encoded diubiquitins to profile linkage selectivity of deubiquitinating enzymes
Nature Communications (2023)